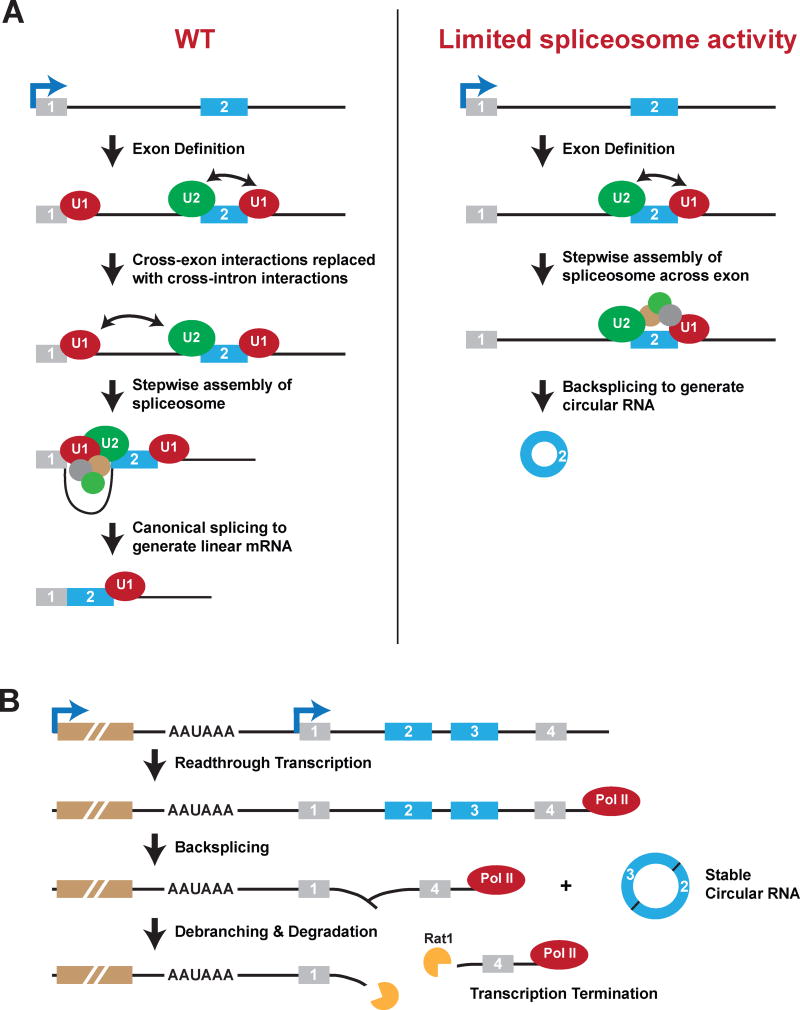
Figure 7. Proposed models for how inhibition or slowing of canonical pre-mRNA processing events can result in increased circular RNA levels.
(A) In wild-type (WT) cells (left), exons within pre-mRNAs are first defined and spliceosomal components assemble across each exon. U1 snRNP recognizes the downstream 5′ splice site, U2 snRNP binds the upstream polypyrimidine tract and branch point sequence, and factors such as SR proteins mediate cross-exon interactions. These cross-exon interactions are subsequently replaced with cross-intron interactions to enable full assembly of the spliceosome and generation of a linear mRNA. When spliceosome activity is limiting (e.g. due to depletion of core spliceosome components) (right), we propose that cross-exon interactions are not easily replaced with cross-intron interactions. The full spliceosome thus assembles across an exon, resulting in backsplicing and the generation of a circular RNA. (B) Failure to efficiently terminate transcriptional units (e.g. due to depletion of cleavage/polyadenylation factors) can cause nascent RNAs to be extended into downstream genes. If the appropriate signals are present in this readthrough transcript (such as inverted intronic repeats), backsplicing can occur to release a mature circular RNA. The remaining nascent RNA likely consists of a Y-shaped structure with a 2′-5′ phosphodiester bond at the upstream branch site. This structure may be debranched, thereby providing an entry site for exonucleases, including Rat1, that enable transcription termination.