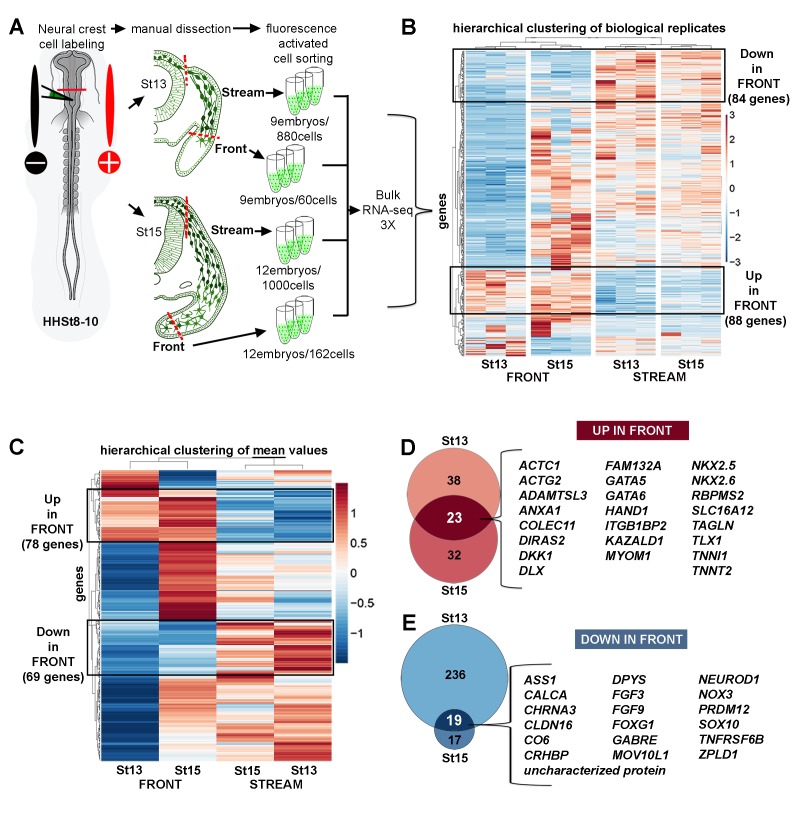
Figure 1. Bulk RNA-seq confirms the unique molecular profile of cells at the invasive front of the neural crest stream.
(A) Schematic representation of method used for harvesting samples from the cranial NC stream. Invasive front (Front) is the ventral-most 5% of NC cells. Stream is the remaining 95% of NC cells. n values as shown. (B) Heatmap of biological replicates of differentially expressed genes in HHSt13 and 15 Front and Stream (n = 477 genes). Black boxes contain gene clusters expressed at significantly higher and lower levels by the Front. (C) Heatmap of mean values of differentially expressed genes in HHSt13 and 15 Front and Stream samples (n = 477 genes). Black boxes contain gene clusters expressed at significantly higher and lower levels by the Front. (D) Venn diagram displaying the numbers of genes enriched at the Front of the NC stream at both HHSt13 and 15 with a union of 23 genes enriched in both comparisons. (E) Venn diagram displaying the numbers of genes reduced at the front of the NC stream at both HHSt13 and 15 with a union of 19 genes reduced in both comparisons. Hamburger and Hamilton (1951); St, stage.