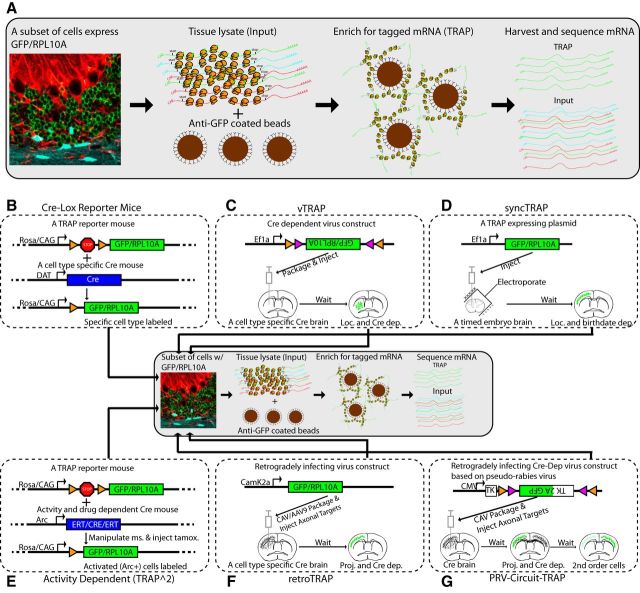
Figure 1.
Alternative methods for targeting cell types for TRAP. A, Illustration of standard TRAP work flow. A specific population of cells is driven to express a GFP-ribosomal fusion protein (GFP/RPL10A, green) that allows for immunofluorescence to confirm targeting and provides a tag for affinity purification of ribosomes from the targeted cell types by using anti-GFP magnetic beads. Initially, cells were targeted using bacterial artificial chromosomes with known expression patterns (Table 1). B, A variety of mouse lines (Table 2) now allow GFP/RPL10A or related construct expression in response to the presence of Cre recombinase, which removes a stop cassette flanked by LoxP sites. This allows profiling of the cell types from a large number of existing Cre mouse lines. C, Use of Cre-dependent virus constructs (here illustrated as the double-inverted operon or FLEX cassette, which flips into forward orientation in the presence of Cre), allows access to Cre-expressing cells in specific regions of the adult brain. D, GFP/RPL10A can also be electroporated into the brain at a defined point in development to label a synchronized population of cells born in a particular window. E, Use of a GFP/RPL10A reporter mouse and an activity-dependent inducible Cre allows for profiling of cells activated by a specific behavior. F, Use of a generic neuronal promoter and a retrogradely infecting virus can be used to profile cells based on their projections. G, Coupling with transsynaptic viruses allows for profiling of second-order (or greater) neurons for a given region and/or cell type.