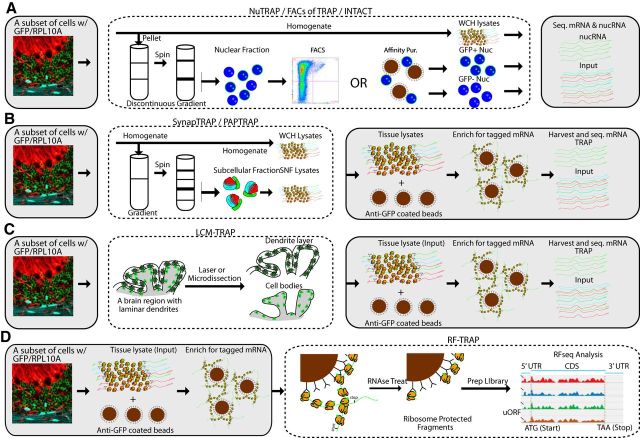
Figure 3.
Adaptations of TRAP for studying subcellular RNA localization and regulation of translation. Additional steps can be inserted into the standard TRAP workflow (gray boxes) to enable new investigations. A, GFP/RPL10A is found in the exterior of nuclear membrane (as it is contiguous with the endoplasmic reticulum) and in the nucleoli. Thus, a simple nuclear fraction can be flow-sorted to harvest cell type-specific nuclei for either nuclear RNAseq or epigenetic studies. It is possible that direct affinity purification for GFP might be possible, as has been shown for other nuclear tags in NuTRAP and INTACT. B, Tissue can be prefractionated using classic protocols into specific subcellular compartments based on density to harvest portions of cells enriched in particular membrane-enclosed fragments, such as synaptoneurosome-containing fractions (SNF), which also contain markers of peripheral astrocyte processes. The fractions can then be lysed, and cell type-specific, subcellularly localized ribosomes can be captured with TRAP. C, Likewise, for those cells where dendrites are in lamina that are physically separable from cell bodies, microdissection enables TRAP on each layer independently. D, TRAP'ed ribosomes can be subjected to RNase treatment to leave only those ∼30 nucleotide fragments physically protected by the ribosome. Sequencing these “ribosome footprints” (RF) allows for analysis of use of specific ORFs from each mRNA.