Fig. 8.
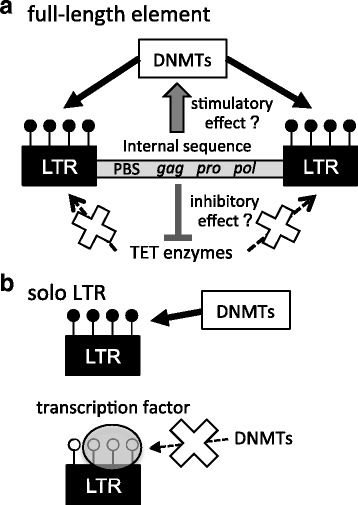
Models for molecular mechanisms that regulate IAP methylation. a In copies containing the internal sequence, some parts of the internal sequence may stimulate DNA methylation in their flanking LTR sequences. Closed circles represent methylated CpG sites in LTRs. DNMTs, DNA methyltransferases. TET enzymes, Ten-Eleven translocation enzymes (catalyzing oxidation of methylcytosine). PBS, primer binding site. b Most solo LTR copies are methylated by DNMTs (top). However, copies carrying TFBSs bind to these TFs if the TFs are expressed in the cell. This binding inhibits the action of DNMTs (bottom). The bound transcription factor is represented as a gray oval. Open circles represent unmethylated CpG sites in the LTRs
