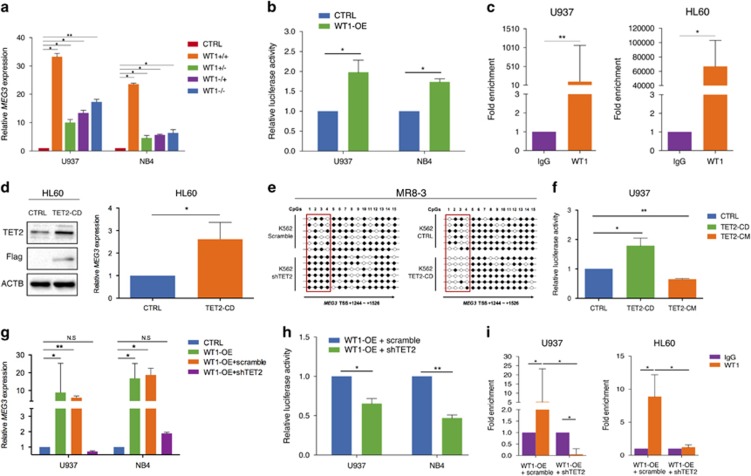
Figure 4.
WT1 interacts with TET2 to co-regulate MEG3 expression in AML cells. (a) RT-qPCR analysis of MEG3 RNA expression after transfected with four major WT1 splicing variants (+5/+KTS, +5/−KTS, −5/+KTS, −5/−KTS) in U937 and NB4 cell lines. (b) Induction of MEG3 promoter activity by WT1 in U937 and NB4 cell lines. (c) The WT1 binding at the promoter regions of MEG3 was assessed by ChIP analysis. (d) HL-60 cell line was transfected with a Flag-TET2CD construct, western blotting analysis of TET2 and Flag, RT-qPCR analysis of MEG3 RNA expression. (e) A representative methylation pattern of the CpGs in K562 cells after bisulfite treatment. Each line represented one PCR product, and six PCR products were shown for each sample. (f) Induction of MEG3 promoter activity by TET2CD or TET2CM in U937 cells. (g) WT1 was transiently overexpressed either singly or with shRNA against TET2 in U937 and NB4 cells, RT-qPCR was examined for the RNA expression of MEG3. (h) Induction of MEG3 promoter activity by WT1 either singly or with shRNA against TET2 in U937 and NB4 cell lines. (i) WT1 was transiently overexpressed either singly or shRNA against TET2 in U937 and HL-60 cells. The WT1 binding at the promoter regions of MEG3 was assessed by ChIP analysis. Representative images of three independent experiments were shown. ACTB protein was used as an internal control for western blotting analysis.*P<0.05; **P<0.01. •, methylated CpG; ○, unmethylated CpG; ChIP, chromatin immunoprecipitation; NS, not significant; RT-qPCR, real-time quantitative PCR; shRNA, short hairpin RNA.