Abstract
An international survey of house dust collected from eleven countries using a modified dilution-to-extinction method yielded 7904 isolates. Of these, six strains morphologically resembled the asexual morphs of Aureobasidium and Hormonema (sexual morphs ?Sydowia), but were phylogenetically distinct. A 28S rDNA phylogeny resolved strains as a distinct clade in Dothideales with families Aureobasidiaceae and Dothideaceae their closest relatives. Further analyses based on the ITS rDNA region, β-tubulin, 28S rDNA, and RNA polymerase II second largest subunit confirmed the distinct status of this clade and divided strains among two consistent subclades. As a result, we introduce a new genus and two new species as Zalaria alba and Z. obscura, and a new family to accommodate them in Dothideales. Zalaria is a black yeast-like fungus, grows restrictedly and produces conidiogenous cells with holoblastic synchronous or percurrent conidiation. Zalaria microscopically closely resembles Hormonema by having only one to two loci per conidiogenous cell, but species of our new genus generally has more restricted growth. Comparing the two species, Z. obscura grows faster on lower water activity (aw) media and produces much darker colonies than Z. alba after 7 d. Their sexual states, if extant, are unknown.
Keywords: 18S, 28S, BenA, black yeast, Dothidiomycetes, ITS, RPB2, xerotolerant fungi, Zalaria
INTRODUCTION
The average person in industrialized countries spends approximately 90 % of their time indoors (Höppe & Martinac 1998). This makes the indoor environment one of the most important human-fungal interfaces. We are constantly exposed to fungal spores, fragments, and metabolites and their impact ranges from human health (Piecková & Jesenská 1999) as pathogens (De Hoog et al. 2014, Garber 2001) or allergens (Aimanianda et al. 2009, Karvala et al. 2011, Tanno et al. 2016) to food spoilage (Pitt & Hocking 2009, Samson et al. 2010) or damage to building materials (Flannigan & Miller 2011). Although indoor environments are not generally recognized as extreme environments, microclimates such as dishwashers contain relatively high water activity (aw) coupled with high temperatures (Zalar et al. 2011), and building materials like plaster, drywall, and cement have very low aw (Flannigan & Miller 2011), while plastics like polyvinyl chloride (PVC, used in construction) offer little in the way of available carbon but still support oligotrophic fungi (Webb et al. 2000). Studying the indoor mycobiota is therefore important to better understand these interactions and how they may affect us.
The black yeast Aureobasidium pullulans (Dothideales) is recorded from a wide variety of sources, including environments with significant osmotic stress, such as hypersaline waters in salterns (Gunde-Cimerman et al. 2000), bathrooms, food, and feeds (Samson et al. 2010), water-damaged wood (Andersen et al. 2011), and polythermal glaciers (Zalar et al. 2008). In surveys of the indoor environment, A. pullulans is one of the most abundant and widespread fungi reported (Adams et al. 2013, Amend et al. 2010, Nonneman et al. 2012, Van Nieuwenhuijzen et al. 2016). The morphospecies exhibits a high degree of phenotypic plasticity (Slepecky & Starmer 2009) and strains can have significantly different pigmentation (Yurlova et al. 1995, Zalar et al. 2008). While this high degree of variation may contribute to its unique adaptability (Gostinčar et al. 2014), it also makes definitive morphological identification challenging, and the many ITS variants identified as this species in GenBank are unlikely to represent one species. Reports of A. pullulans being one of the most abundant members in fungal communities based on near-neighbour analyses of next-generation sequencing (NGS) data may be skewed to some extent.
The class Dothideomycetes is the largest in Ascomycota and was recently examined and re-defined using multigene phylogenetics (Hyde et al. 2013, Schoch et al. 2009, Thambugala et al. 2014). These studies showed the order Dothideales to be a monophyletic sister to Myriangiales. Thambugala et al. (2014) reviewed and re-evaluated the morphologically-based taxonomy of Dothideales known from culture, informed by a combined phylogeny of 28S rDNA, 18S rDNA and ITS. They accepted two families, synonymising the often-accepted Dothioraceae (e.g. Barr 2001) with Dothideaceae, as first proposed by Von Arx & Müller (1975), and introducing Aureobasidiaceae for Aureobasidium and closely related genera. The polythetic morphological definitions provided by Thambugala et al. (2014) did not identify unique diagnostic characters among the sexual or asexual morphs in either family. Both families include a poorly integrated mixture of genera known from fungarium specimens with others that are mostly known from culture. For both families, sexually and asexually typified genera were keyed out separately. Sexually typified genera were separated by characters of the stromata, number of ascospores per ascus, and ascospore characters such as septation and pigmentation. In Dothideales, the characters normally used to classify asexual morphs were clearly phylogenetically uninformative, and mixtures of black yeast, hyphomycetous, coelomycetous, and intermediate asexual morphs are scattered over the various clades of Aureobasidiaceae and Dothideaceae. Both families include a variety of asexual morphs, including sporodochial hyphomycete forms usually observed in nature (e.g. Kabatiella, Kabatina), coelomycete (e.g. Endoconidioma, Neocylindroseptoria, Rhizosphaera), and black yeast-like forms usually observed in culture.
Black yeasts have a confused taxonomic history that may eventually be clarified with the single name classification system, but presently remains difficult to navigate. Black yeasts are a phylogenetically diverse morphogroup of asexual morphs, mostly in Dothideales or Chaetothyriales, which produce dark, slimy colonies and at least some budding yeast-like cells in culture. Many species have one or more hyphal asexual morphs in addition to the yeast-like forms. This pleiomorphy complicates their identification and taxonomic interpretation (De Hoog & Hermanides-Nijhof 1977). Two of the most frequently reported black yeast genera are Aureobasidium (Aureobasidiaceae) and Hormonema (Dothideaceae). Aureobasidium was associated with several sexually typified genera in Dothideales, and although no sexual morph is definitively known for the most frequently reported species A. pullulans, an ascospore-derived strain identified as Columnosphaeria fagi (CBS 171.93), has identical ITS and RPB2 sequences to the clade including the ex-type culture of A. pullulans. The similar asexually typified genus Hormonema, (H. dematioides type), generally considered the asexual morph of Sydowia polyspora, is also frequently reported. A single name solution for this clade is not yet proposed and we use the name Hormonema for comparisons of asexual morphotypes. Despite the differences in associated sexual morphs, Aureobasidium and Hormonema are difficult to distinguish morphologically when grown in culture. For example, neither Hermanides-Nijhof (1977) nor De Hoog & Yurlova (1994) could find morphological differences among asexual morphs in cultures of the sexually typified genera Pringsheimia, Dothidea, Dothiora or Sydowia, all of which they attributed to Hormonema. Aureobasidium and Hormonema were considered distinct in the dual nomenclature era, because of the different sexual morphs. Hermanides-Nijhof (1977) defined Aureobasidium by the production of synchronous blastoconidia from undifferentiated, hyaline cells, whereas Hormonema was said to produce conidia in basipetal succession from hyaline or dark hyphae. Wang & Zabel (1990) suggested that at least some conidiogenous cells of H. dematioides were phialidic or percurrent. In a later review, Aureobasidium was distinguished based on its conidiogenous cells having multiple loci (synchronous conidiogenesis), in contrast to one or two loci in Hormonema (De Hoog & Yurlova 1994). These characters are difficult to detect, and are best observed along hyphae at the growing margin of the colony. Colonies of both morphs often begin as palely pigmented growths, which become slimy and almost black as the colonies mature. The different patterns and apparent plasticity of conidiogenesis, including yeast-like cells, young blastospores, swollen blastospores, chlamydospores, and septation and constrictions of hyphae confound interpretations of homologous characters (Guterman & Shabtai 1996, Zalar et al. 2008).
During our survey of fungi isolated from house dust using a dilution-to-extinction approach, many isolates morphologically resembled Aureobasidium and Hormonema, but six were phylogenetically distinct. Here we introduce these as two new species, in a new genus and family in Dothideales. We present a 28S rDNA (nuclear large ribosomal subunit) phylogeny of Dothidiomycetes to determine the strains’ phylogenetic position and subsequently present phylogenies of Dothideales based on BenA (β-tubulin), ITS rDNA, 28S rDNA, 18S rDNA (nuclear small ribosomal subunit), and RPB2 (RNA polymerase II second largest subunit), to determine the relationships within the order. Strains were characterized morphologically and compared to morphologically similar species and genera. This work follows previous reports of new taxa of indoor fungi discovered by dilution-to-extinction, including species of Rasamsonia (Tanney & Seifert 2013), Aspergillus, Penicillium and Talaromyces (Visagie et al. 2014, 2017, Sklenář et al. 2017), Wallemia (Jancic et al. 2015, Nguyen et al. 2015), Spiromastix, Pseudospiromastix, Sigleria (Hirooka et al. 2016), and Myrmecridium (Crous et al. 2016).
MATERIAL AND METHODS
Isolations
Settled house dust was collected from twelve countries (Australia, Canada, Federated States of Micronesia, Indonesia, Mexico, The Netherlands, New Zealand, South Africa, Thailand, the United Kingdom, Uruguay, and USA) using sterilized Duststream® collectors (Indoor Biotechnologies, Charlottesville, VA) attached to vacuum cleaners. Isolations were made from malt extract agar (MEA) and MEA with 20 % sucrose using a dilution-to-extinction method modified from Collado et al. (2007) as described in Visagie et al. (2014). More recent isolations targeting xerophilic fungi from Canadian and Hawaiian house dust were made as described in Visagie et al. (2017).
Living strains of new species are deposited in the Canadian Collection of Fungal Cultures (DAOMC, Ottawa, Canada), the Westerdijk Fungal Biodiversity Institute (CBS, Utrecht, The Netherlands) and dried specimens are accessioned in the Canadian National Mycological Herbarium (DAOM, Ottawa, Canada). Strains used in this study are summarized in Table 1.
Table 1.
Information on house dust isolates included in this study.
| Species | Strain number | Isolation medium | Origin | Collectora | Date collected | Isolatorb |
|---|---|---|---|---|---|---|
| Aureobasidium melanogenum | SLOAN 1260 = AA07MX-884 | MEA | Mexico, Nayarit, Sayulita | A. Amend | 31 Jan. 2009 | E. Whitfield & K. Mwange |
| Aureobasidium melanogenum | SLOAN 1606 = BH02AU-110 | MEA | Australia, Tasmania, Hobart | B. Horton | 10 Feb. 2009 | E. Whitfield & K. Mwange |
| Aureobasidium melanogenum | SLOAN 5623 = TA10NZ-214a | MEA | New Zealand, Wellington, Wellington | T. Atkinson | 3 May 2009 | E. Whitfield & K. Mwange |
| Aureobasidium melanogenum | KAS 5840 | MY50G | Canada, Ontario, Stittsville | K.A. Seifert | 20 Dec. 2014 | C.M. Visagie |
| Aureobasidium melanogenum | KAS 7917 | MY1012 | USA, Hawaii, Kailua | A. Amend | 11 May 2015 | C.M. Visagie |
| Aureobasidium melanogenum | KAS 7956 | DG18 | USA, Hawaii, Kailua | A. Amend | 11 May 2015 | C.M. Visagie |
| Aureobasidium melanogenum | SLOAN 7256 = AA04US-587 | 20%S-MEA | USA | A. Amend | unknown | E. Whitfield & K. Mwange |
| Aureobasidium pullulans | SLOAN 7261 = AA02US-306 | 20%S-MEA | USA, California, Berkeley | A. Amend | 31 Mar. 2005 | E. Whitfield & K. Mwange |
| Aureobasidium pullulans | SLOAN 203 = 7050035.79-65 | n.a. | Canada, Saskatchewan, Regina | Health Canada | 12 Mar. 2007 | E. Whitfield & K. Mwange |
| Aureobasidium pullulans | SLOAN 207 = 7050035.79-71 | n.a. | Canada, Saskatchewan, Regina | Health Canada | 12 Mar. 2007 | E. Whitfield & K. Mwange |
| Aureobasidium pullulans | SLOAN 48 = 7050035.79-127 | 20%S-MEA | Canada, Saskatchewan, Regina | Health Canada | 12 Mar. 2007 | E. Whitfield & K. Mwange |
| Aureobasidium pullulans | SLOAN 62 = 7050035.79-148 | 20%S-MEA | Canada, Saskatchewan, Regina | Health Canada | 12 Mar. 2007 | E. Whitfield & K. Mwange |
| Aureobasidium pullulans | SLOAN 349 = 7330009.33-45 | 20%S-MEA | Canada, Saskatchewan, Regina | Health Canada | 21 Aug. 2007 | E. Whitfield & K. Mwange |
| Aureobasidium pullulans | SLOAN 359 = 7330009.33-60 | 20%S-MEA | Canada, Saskatchewan, Regina | Health Canada | 21 Aug. 2007 | E. Whitfield & K. Mwange |
| Aureobasidium pullulans | SLOAN 7249 = 7330009.34-925 | n.a. | Canada, Saskatchewan, Regina | Health Canada | 21 Aug. 2007 | E. Whitfield & K. Mwange |
| Aureobasidium pullulans | SLOAN 1652 = BH02AU-154a | 20%S-MEA | Australia, Tasmania, Hobart | B. Horton | 10 Feb. 2009 | E. Whitfield & K. Mwange |
| Aureobasidium pullulans | SLOAN 1653 = BH02AU-154b | 20%S-MEA | Australia, Tasmania, Hobart | B. Horton | 10 Feb. 2009 | E. Whitfield & K. Mwange |
| Aureobasidium pullulans | SLOAN 3214 = KJ09SA-65 | MEA | South Africa, Western Cape, Kuilsrivier | K. Jacobs | 24 Jul. 2009 | E. Whitfield & K. Mwange |
| Aureobasidium pullulans | KAS 5951 | DG18 | Canada, British Columbia, Victoria | B. Kendrick | 27 Jan. 2015 | C.M. Visagie |
| Aureobasidium species | SLOAN 41 = 7050035.79-119 | MEA | Canada, Saskatchewan, Regina | Health Canada | 12 Mar. 2007 | E. Whitfield & K. Mwange |
| Aureobasidium subglaciale | SLOAN 7263 = AA02US-332 | 20%S-MEA | USA, California, Berkeley | A. Amend | 31 Mar. 2005 | E. Whitfield & K. Mwange |
| Hortaea werneckii | KAS 7942 | MY50G | USA, Hawaii, Kailua | A. Amend | 11 May 2015 | C.M. Visagie |
| Hortaea werneckii | KAS 7947 | MY50G | USA, Hawaii, Kailua | A. Amend | 11 May 2015 | C.M. Visagie |
| Hortaea werneckii | KAS 7949 | MY1012 | USA, Hawaii, Kailua | A. Amend | 11 May 2015 | C.M. Visagie |
| Hortaea werneckii | KAS 7953 | MY1012 | USA, Hawaii, Kailua | A. Amend | 11 May 2015 | C.M. Visagie |
| Rhizosphaera pini | DAOMC 251499 = KAS 6309 | MY50G | Canada, New Brusnwick, Little Lepreau | A. Walker | 29 Jan. 2015 | C.M. Visagie |
| Sydowia polyspora | DAOMC 251470 = KAS 5918 | DG18 | Canada, British Columbia, Victoria | B. Kendrick | 27 Jan. 2015 | C.M. Visagie |
| Sydowia polyspora | DAOMC 251471 = KAS 5919 | DG18 | Canada, British Columbia, Victoria | B. Kendrick | 27 Jan. 2015 | C.M. Visagie |
| Sydowia polyspora | DAOMC 251469 = KAS 5999 | DG18 | Canada, New Brusnwick, Little Lepreau | A. Walker | 29 Jan. 2015 | C.M. Visagie |
| Zalaria alba | DAOMC 250847 = SLOAN 52 = 7050035-79-132 | 20%S-MEA | Canada, Saskatchewan, Regina | Health Canada | 12 Mar. 2007 | E. Whitfield & K. Mwange |
| Zalaria alba | DAOMC 250848 = SLOAN 352 = 7330009.33-5 | MEA | Canada, Saskatchewan, Regina | Health Canada | 21 Aug. 2007 | E. Whitfield & K. Mwange |
| Zalaria obscurum | DAOMC 250851 = SLOAN 7266 = AA02US-340 | 20%S-MEA | USA, California, Berkeley | A. Amend | 31 Mar. 2005 | E. Whitfield & K. Mwange |
| Zalaria obscurum | DAOMC 250852 = SLOAN 7674 = AA02US-351 | 20%S-MEA | USA, California, Berkeley | A. Amend | 31 Mar. 2005 | E. Whitfield & K. Mwange |
| Zalaria obscurum | DAOMC 250849 = SLOAN 7244 = 7330009.34-884 | n.a. | Canada, Saskatchewan, Regina | Health Canada | 21 Aug. 2007 | E. Whitfield & K. Mwange |
| Zalaria obscurum | DAOMC 250850 = SLOAN 7246 = 7330009.34-921 | n.a. | Canada, Saskatchewan, Regina | Health Canada | 21 Aug. 2007 | E. Whitfield & K. Mwange |
a Collector of house dust sample; b isolator of strain using the modified dilution to exctinction method.
Morphology
The strains considered here were suspected to be xerophilic and were thus characterized from colonies grown on a wide range of media, including MEA, potato-dextrose agar (PDA; Oxoid CM139), oatmeal agar (OA), dichloran 18 % glycerol agar (DG18; Hocking & Pitt 1980), starch-nitrate agar (SNA; Dodman & Reinke 1982), malt extract yeast extract with 50 % glucose agar (MY50G), malt extract yeast extract 10 % glucose 12 % NaCl agar (MY10-12; Pitt & Hocking 2009), and MEA with the addition of 5–24 % NaCl (MEA5NaCl, MEA10NaCl, MEA15NaCl, MEA24NaCl). The malt extract used for media was always BD BactoTM (Mississauga, ON). Plates were incubated for 7 and 14 d in the dark at 25 °C. Additional MEA plates were incubated at 10 and 30 °C. Colour names and codes used in descriptions refer to Kornerup & Wanscher (1967). Microscope preparations were made from colonies grown on MEA and DG18, using lactic acid as mounting fluid. An Olympus BX50 compound microscope attached with an InfinityX camera powered by Infinity Analyze v. 6.5.1 software (Lumenera, Ottawa, ON) was used for microscopic observations, capturing images and making measurements. Photographic plates were prepared in Affinity Photo v. 1.5.2 (https://affinity.serif.com).
DNA extraction, sequencing, and analysis
DNA was extracted from 7–10-day-old cultures grown on Blakeslee’s malt extract agar (MEA; (Blakeslee 1915)) using the UltraClean™ Microbial DNA isolation Kit (MoBio Laboratories, Solano Beach, CA) with extracts stored at −20 °C. Loci were amplified using the following primer pairs: 28S rDNA with LR0R & LR5 (Vilgalys & Hester 1990); ITS with V9G/LS266 (Gerrits van den Ende & De Hoog 1999, Masclaux et al. 1995); RPB2 with fRPB2-5F/ fRPB2-7cR (Liu et al. 1999); 18S rDNA with NS1/NS4 (White et al. 1990); and BenA with T10/Bt2b (Glass & Donaldson 1995, O’Donnell & Cigelnik 1997). An annealing temperature of 55 °C was used for all reactions. PCR amplification was performed in 10 μL volume reactions, containing 0.5 μL template DNA, 1 μL Titanium Taq buffer (Takara Bio USA, Mountain View, CA), 0.5 μL (2 mM) dNTP’s, 0.04 μL (3.2 mM) of each primer, 0.1 μL Titanium Taq polymerase (Takara Bio USA), and 7.82 μL MilliQ water.
Sequencing reactions were set up using the BigDye™ Terminator v. 3.1 Cycle Sequencing Kit (Applied Biosystems, Waltham, MA) and the same primer pairs used for PCR amplification, with additional sequence reactions set up for 28S rDNA with primers LR3/LR3R (Vilgalys & Hester 1990). Sequence contigs were assembled in Geneious v. 8.1.8 (BioMatters, Auckland, NZ) and are deposited in GenBank (KX579092–KX579121, KY654326, KY659498, KY659500–KY659528). Accession numbers are also displayed on phylogenetic trees. BLAST searches were performed using NCBI to determine closest sequence matches and whether our species were detected in previous studies.
Phylogenetic analyses
The phylogenetic position of our strains within Dothideomycetes was determined using a 28S rDNA phylogeny compared to reference sequences obtained from Schoch et al. (2009) and Hyde et al. (2013). Secondly, phylogenies of BenA, ITS, 28S rDNA, RPB2 and 18S rDNA were used to determine the relationships among our strains within the new genus, and the relationship of the genus and family with close relatives in Dothideales and Myriangiales. Newly generated sequences obtained from dust isolates belonging to Aureobasidium, Hortaea, Rhizosphaera, and Sydowia are also included in the phylogenies. Reference sequences for comparisons were obtained from GenBank and accession numbers are included on trees.
Datasets were aligned using MAFFT v. 7.017 (Katoh & Standley 2013) with the L-INS-i algorithm. For the BenA dataset, G-INS-i was used. Alignments were manually trimmed in Geneious. The Dothideomycetes phylogeny was calculated using RAxML v. 8.0.0 (Stamatakis 2014) and support at nodes calculated using a bootstrap analysis of 1000 replicates. Additional phylogenies were calculated based on Maximum Likelihood done in RAxML and Bayesian tree interference (BI) using MrBayes v. 3.2 (Ronquist et al. 2012). For BI, the most suitable model for each dataset was determined using MrModeltest v. 2.3 (Nylander 2004) based on the lowest Akaike information criteria (Akaike 1974) value. Trees were visualized in Figtree v. 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree) and prepared for publication in Adobe® Illustrator® CS6. Aligned datasets and trees were uploaded to TreeBASE (www.treebase.org) with submission ID 19764.
RESULTS
Phylogeny
Dothideomycetes 28S rDNA phylogeny (Fig. 1): The aligned 28S rDNA dataset contained 515 taxa and was 2448 bp long. Schismatomma decolorans was selected as outgroup based on Schoch et al. (2009). In general, our phylogeny shared similar topologies to those observed in Schoch et al. (2009) and Hyde et al. (2013). The phylogeny placed our isolates within the monophyletic order Dothideales, although there was poor support on the branch separating it from Myriangiales. Furthermore, our isolates resolved in a clade distinct from the two families currently recognized in Dothideales, i.e. Aureobasidiaceae and Dothideaceae. Therefore, we introduce the new genus Zalaria and classify it in a new family named Zalariaceae below.
Fig. 1.
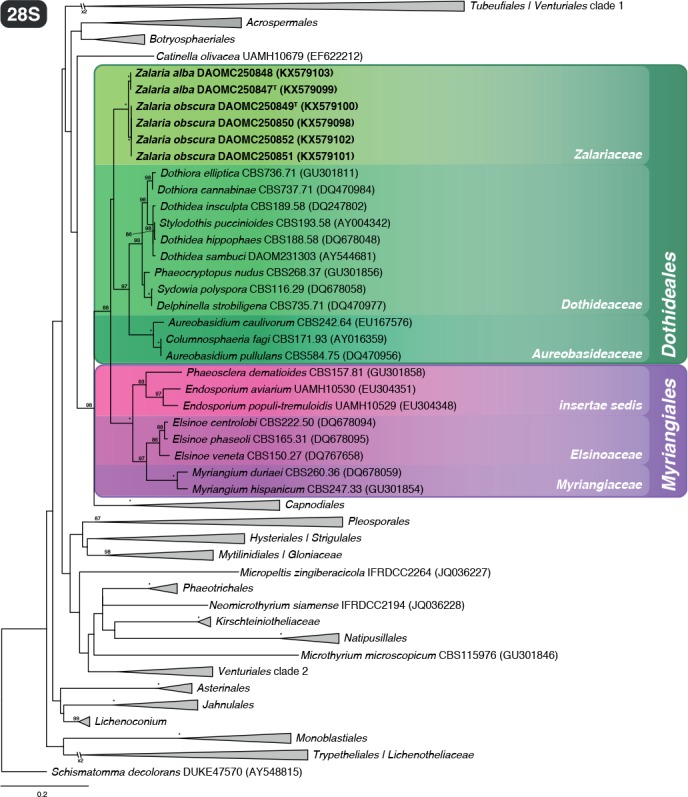
Phylogenetic tree of Dothideomycetes based on 28S rDNA showing the distinct nature of the new family Zalariaceae within Dothideales. Schismatomma decolorans was selected as outgroup. Bootstrap support values higher than 80 % are indicated above branches (* indicates 100 % bootstrap support). House dust isolates are shown in bold text.
Dothideales phylogenies (Figs 2–3): To examine relationships and the phylogenetic species concept within Zalaria and its relationship with related families more closely, we calculated focused phylogenies based on 18S rDNA, 28S rDNA, BenA, ITS, and RPB2, including only closely related species and genera from Dothideaceae and Aureobasidiaceae, as well as species in the order Myriangiales. All loci consistently resolved the Zalaria strains in clades distinct from Aureobasidiaceae and Dothideaceae. In the ITS phylogeny, they resolved as a close relative of Myriangiales, but this tree had poor backbone support. All loci, except for the highly conserved 18S rDNA, resolved strains into two clades, which were strongly supported in 28S rDNA, BenA, ITS, and RPB2. They are described below as Zalaria alba and Z. obscura spp. nov. Furthermore, the 28S rDNA phylogeny revealed several strains that we consider identical to Z. obscura. These strains (CBS 122350; CBS 122359; EXF-922; EXF-1934; EXF-1936) originated from Norwegian arctic ice and were published in Zalar et al. (2008), but were never given a formal name. Remaining house dust isolates were identified here as Aureobasidium pullulans, A. melanogenum, A. subglaciale, an undescribed Aureobasidium species (DTO 285-D8), Rhizosphaera pini, Sydowia polyspora, and Hortaea werneckii (Capnodiales).
Fig. 2.
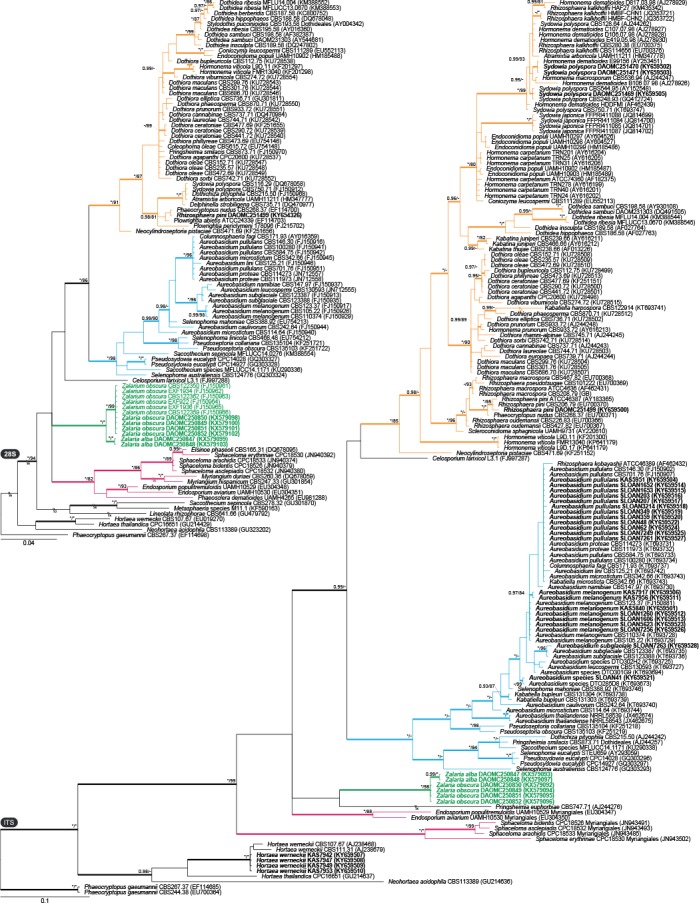
Phylogenies based on 28S rDNA and ITS showing the relationship of Zalaria (green text and branches) with other closely related genera from families Dothideaceae (orange branches), Aureobasidiaceae (blue branches) and order Myriangiales (maroon branches). Bootstrap support values or Bayesian posterior probabilities higher than 79 % or 0.94 are indicated above thickened branches (* indicates 100 % or 1.00; – indicates lack of support). House dust isolates from this study are indicated by bold text. GenBank accession numbers are provided between brackets.
Fig. 3.
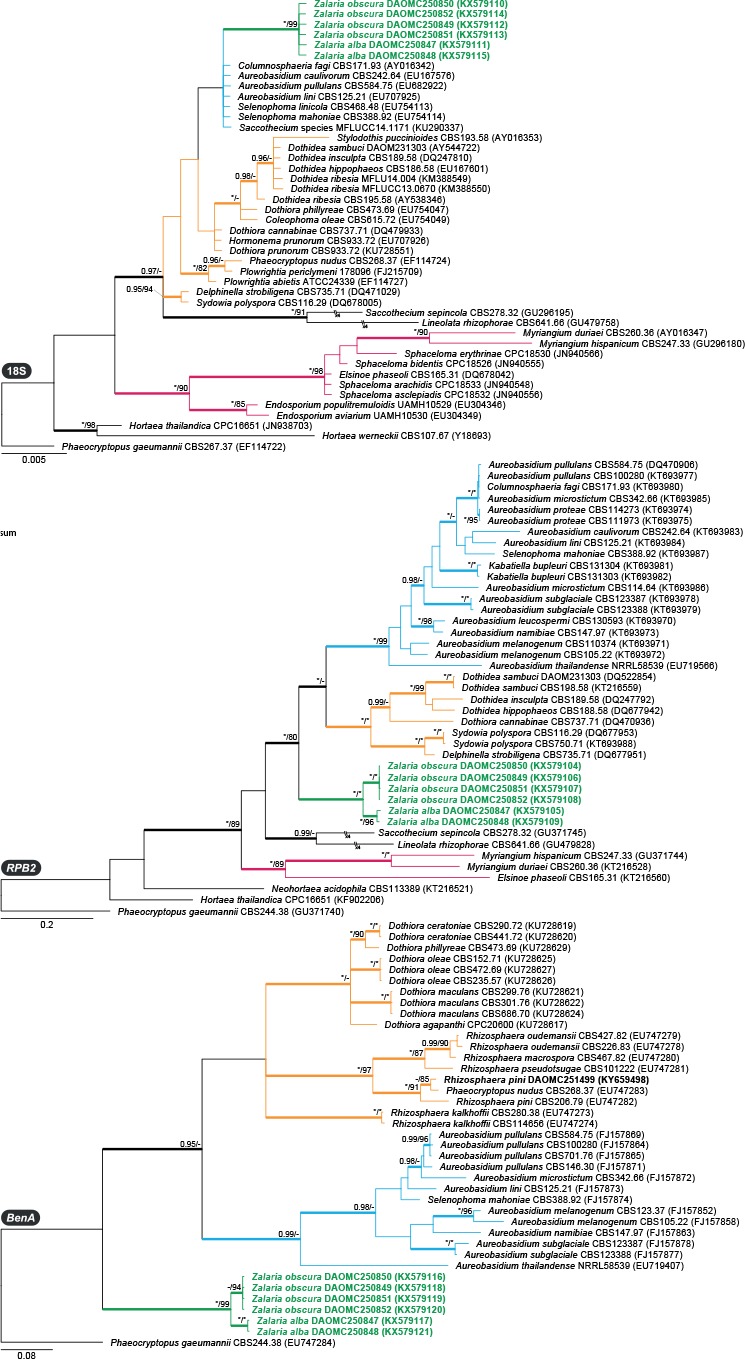
Phylogenies based on 18S rDNA, RPB2 and BenA showing the relationship of Zalaria (green text and branches) with other closely related genera from families Dothideaceae (orange branches), Aureobasidiaceae (blue branches) and order Myriangiales (maroon branches). Bootstrap support values or Bayesian posterior probabilities higher than 79 % or 0.94 are indicated above thickened branches (* indicates 100 % or 1.00; – indicates lack of support). House dust isolates from this study are indicated by bold text. GenBank accession numbers are provided between brackets.
NCBI-BLAST (Table 2): BLAST searches resulted in several hits similar to Zalaria ITS and 28S rDNA sequences. These sequences were from a diverse range of studies and originate from the USA, China, Greece, The Netherlands, Portugal, Spain, Thailand, and Antarctica and from habitats including house dust, cork samples, grapes, a hospital incubator, sediment, soil, wood, and woodpecker excavations. Our Zalaria isolates were obtained from house dust collected in the USA (CA) and Canada (Regina, SK).
Table 2.
BLAST results showing origin of strains or clones belonging to Zalaria.
| Gene | GenBank nr | Original identification | Current identification | Strain/clone | Origin and source | Publication |
|---|---|---|---|---|---|---|
| ITS | KM103983 | Uncultured fungus | Zalaria cf sp2 | Rcw 47 | USA; Red-cockaded woodpecker excavation in living pine | Unpublished |
| ITS | KJ130063 | Aureobasidium sp. | Zalaria cf sp2 | C 21 | China, Henan, Nanyang City, Baotianman Natural Reserve Area; forest soil | Unpublished |
| ITS | KF800358 | Uncultured fungus | Zalaria cf sp2 | CMH 267 | USA, Missouri, Kansas City; House dust | Rittenour et al. (2014) |
| ITS | KC253970 | Aureobasidium pullulans | Zalaria cf sp2 | UOA/HCPF ENV57A | Greece; hospital incubator | Unpublished |
| ITS | FJ235939 | Fungal sp. | Zalaria sp1 | AB 6 | Antarctica; wood | Unpublished |
| ITS | AF121282 | Aureobasidium pullulans | Zalaria sp2 | ATCC 16628 | The Netherlands; Soil | Unpublished |
| LSU | AB456556 | Aureobasidium sp. | Zalaria cf sp2 | WB 27 | Thailand, Prachuap Khiri Khan, Pran Buri Forest Park; sediment in mangrove forest | Unpublished |
| LSU | AY167611 | Aureobasidium sp. | Zalaria cf sp2 | CECT 11965 | Spain?; cork samples | Álvarez-Rodríguez et al. (2003) |
| LSU | KJ130062 | Aureobasidium sp. | Zalaria cf sp2 | C 21 | China, Henan, Nanyang City, Baotianman Natural Reserve Area; forest soil | Unpublished |
| LSU | JN004186 | Aureobasidium sp. | Zalaria cf sp2 | SP 9 | Portugal; Sound grapes | Unpublished |
| LSU | KC433818 | Aureobasidium sp. | Zalaria cf sp2 | DBVPG 5996 | Italy, Mont Blanc, Miage glacier; supraglacial sediments | Turchetti et al. (2013) |
Morphology
Strains were characterized morphologically on several agar media and shared several characters with species of Aureobasidium and Hormonema. As noted in the Introduction, the only way to reliably distinguish between these groups are the 1–2 conidiogenous loci per cell in Hormonema (and asexual morphs of many other Dothideaceae species), and up to 14 loci in Aureobasidium (De Hoog & Yurlova 1994, Yurlova et al. 1999). The new genus is more similar in this regard to Hormonema, with only 1–2 loci per conidiogenous cell. In general, growth of Zalaria species is more restricted than in any of these other genera.
Three types of conidiogenesis were observed in the Zalaria strains. Their yeast forms are very common, especially in younger colonies when cells reproduce by budding. After prolonged incubation, these yeast cells are often covered by melanized hyphal growth; microscopic observations suggest that hyphae from germinating yeast cells eventually give rise to this dark melanized growth (Figs 4C–D, 5C–D). With age, these hyphae may develop into dark brown, thick-walled chlamydospores (Figs 4E, I, 5E, I). Further, intercalary conidiogenous cells develop mostly at margins of young colonies.
Fig. 4.
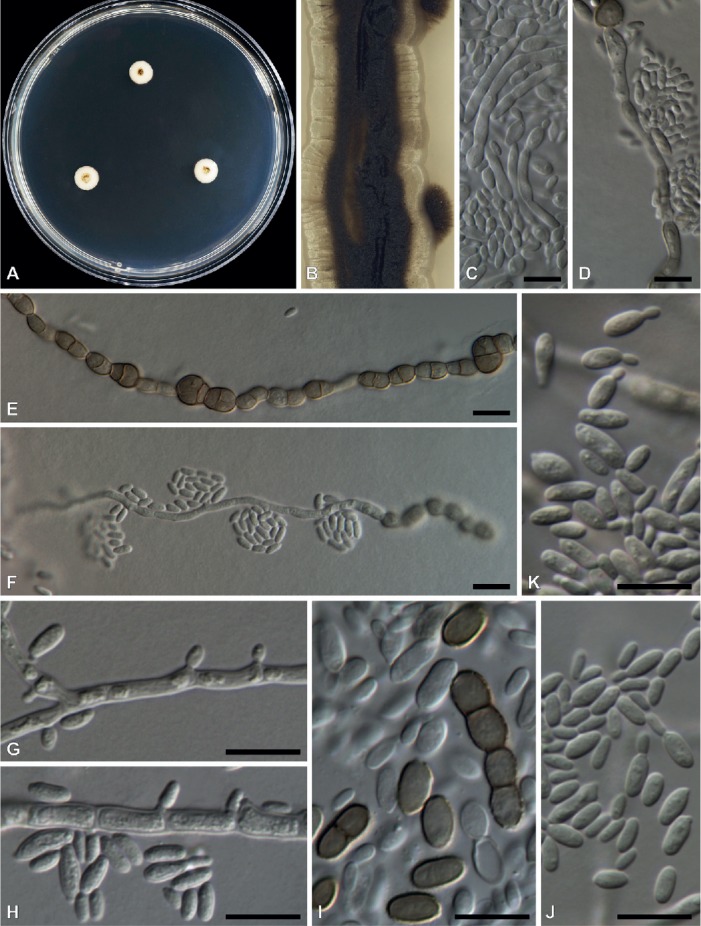
Morphological characters of Zalaria alba (DAOMC 250847 in A–C, E, F; DAOMC 250848 in D, G–K). A. Colonies on MEA after 2 wk. B. Close-up colony on MEA after 4 wk. C. Germinating conidia. D. Germinating conidia with age. E. Melanized hyphae developing into arthrospores and chlamydospores. F–H. Intercalary conidiogenous cells. I. Chlamydospores. J–K. Conidia with some yeast-like budding occurring. Bars = 10 μm.
Fig. 5.
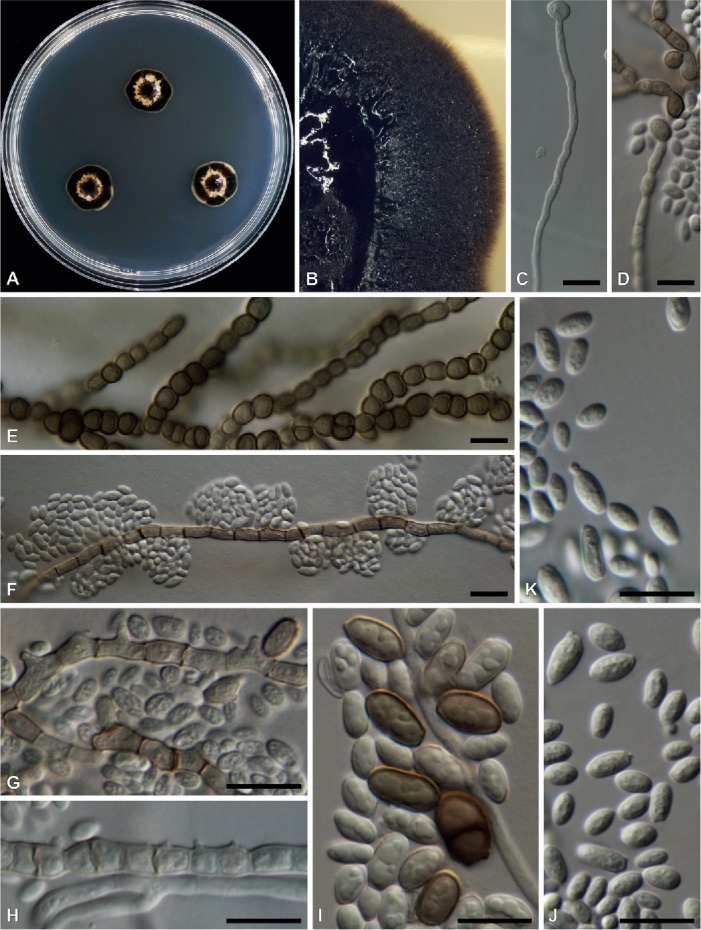
Morphological characters of Zalaria obscura (DAOMC 250849 in A, B, D, F, G, J, K; DAOMC 250852 in C, H, I; DAOMC 250850 in E). A. Colonies on MEA after 2 wk. B. Close-up colony on MEA after 4 wk. C. Germinating conidia. D. Germinating conidia with age. E. Melanized hyphae developing into chlamydospores. F–H. Intercalary conidiogenous cells. I. Chlamydospores. J–K. Conidia with some yeast-like budding occurring. Bars = 10 μm.
The strains resolved as distinct clades observed in the phylogenies were also distinct morphologically, with Z. alba compared to Z. obscura growing more restrictedly on most media. Zalaria obscura is also capable of growth at low aw media such as MEA-10%-NaCl, MY-1012 and MY50G, with Z. alba not growing on these media. Generally, Z. alba colonies are also more yeast-like and take up to 3 wk to darken, whereas Z. obscura colonies darken in 7 d and are often covered by a leathery layer within 14 d. This character was originally used for distinguishing the two varieties of A. pullulans var. pullulans and var. melanogenum, later recognized as two distinct species (Gostinčar et al 2014). Based on both phylogenetic and morphological results we introduce a new family, genus and two new species to accommodate our unique black yeast-like fungi.
Morphological examinations confirmed sequence-based identifications of the remaining house dust isolates (Figs 6–7). Aureobasidium strains produced the typical conidiogenous cells with multiple loci (Fig. 6A–G), while only 1–2 conidiogenous loci were observed in strains identified as Sydowia polyspora (Fig. 6H–K). Hortaea werneckii was common in Hawaiian dust samples and was only isolated from the halophilic MY10-12 medium. The typical pigmented hyphae, yeast-like growth, sympodial and percurrent conidiogenesis were observed in newly isolated strains (Fig. 7A–E). The strain identified as Rhizosphaera pini produced colonies with pycnidium-like structures and a Hormonema-like morph producing very large conidia, all characteristic of that species (Fig. 7F–I).
Fig. 6.
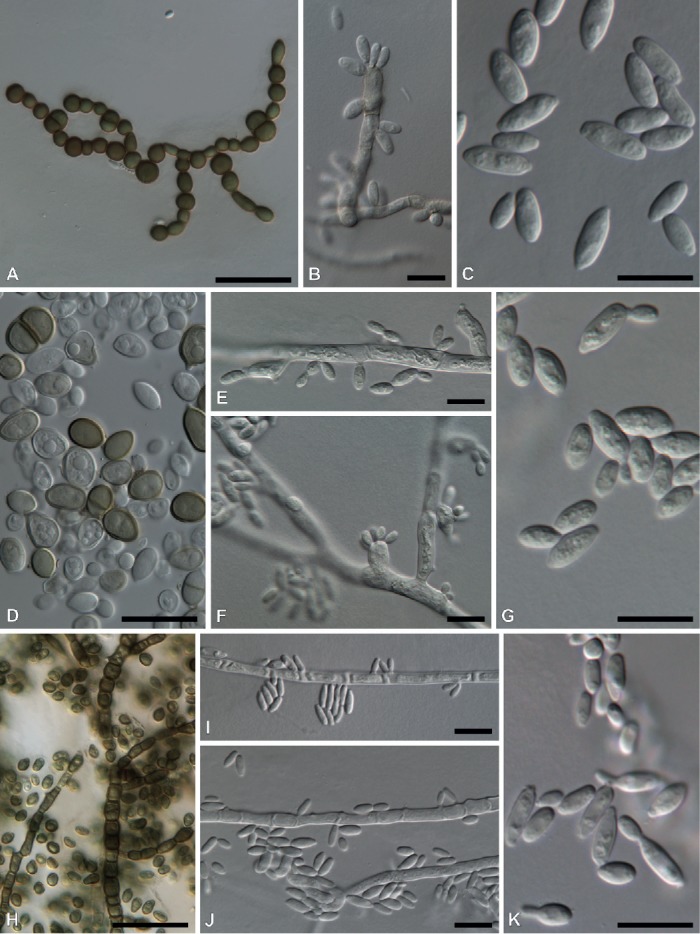
A–C. Aureobasidium pullulans (KAS 5840). A. Melanized hyphae/chlamydospores. B. Conidiogenous cells with multiple loci. C. Conidia. D–G. Aureobasidium melanogenum (KAS 1951). D. Dark brown conidia. E–F. Conidiogenous cells with multiple loci. G. Conidia. H–K. Sydowia polyspora (DAOMC 251471). H. Melanized hyphae/chlamydospores. I–J. Hormonema-like conidiogenous cells with 1–2 loci. K. Conidia. Bars = 10 μm except A and H = 50 μm.
Fig. 7.
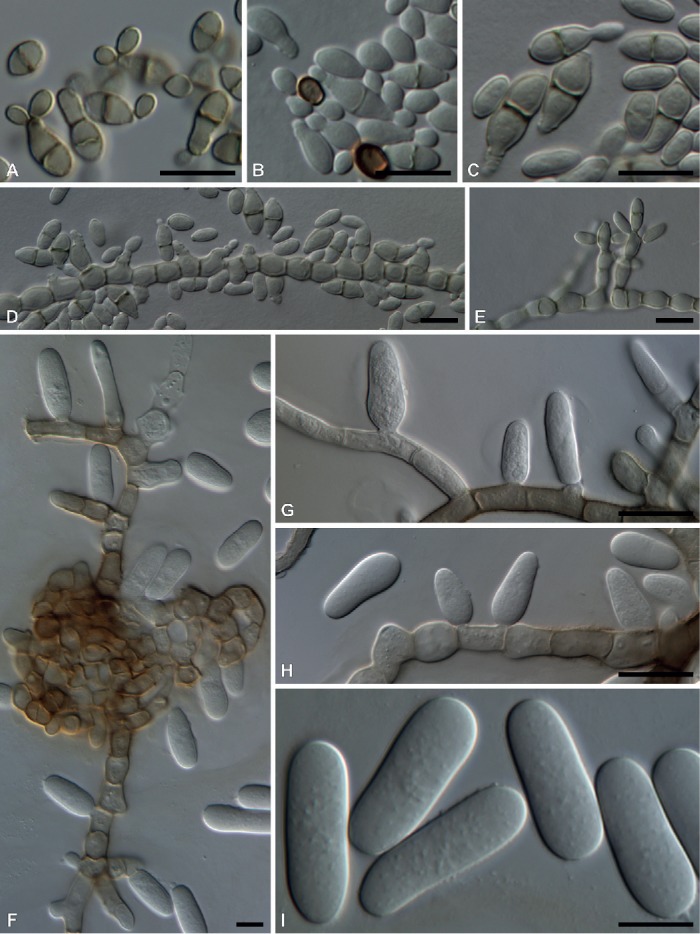
A–E. Hortaea werneckii (DAOMC 251499). A. Yeast-like cells with sympodial producing daughter cells. B–C. Yeast-like cell with annellations. D–E. Conidiogenous apparatus. F–I. Rhizosphaera pini (DAOMC 251499). F–H. Hormonema-like conidiogenous cells. I. Conidia. Bars = 10 μm.
Taxonomy
Zalariaceae Visagie, Z. Humphries & Seifert, fam. nov.
MycoBank MB821627
Type genus: Zalaria Visagie et al. 2017.
Diagnosis: Distinguished from other families classified in Dothideales and Myriangiales based on a short unique 28S rDNA sequence flanked by two conserved regions. The section defining Zalariaceae in our alignment (Treebase ID 19764) are found between nucleotide positions 39 to 62 and is indicated in bold text (5’-AGCTCAAATTTGAAATCTGGCC-CTTTC-AGGGTCCGAGTTGTAATTTGTAGAGG-3’).
Zalaria Visagie, Z. Humphries & Seifert, gen. nov.
MycoBank MB821628
Etymology: Named in honour of Polona Zalar, mycologist at the University of Ljubljana, Slovenia, in recognition of her studies on extremophilic fungi, including her 2008 study that included strains from Norwegian arctic regions that belong to this genus.
Diagnosis: Differs from Aureobasidium by blastic conidiogenesis occurring from one to two loci per conidiogenous cell. Morphologically Zalaria is indistinguishable from Hormonema (often reported as asexual morphs of Sydowia), leaving DNA sequences the only diagnostic character (see Diagnosis for family Zalariaceae above).
Type species: Zalaria obscura Visagie et al. 2017
Description: Sexual morph unknown. Colonies often covered in slimy masses of conidia or yeast-like cells, becoming dark and often leathery with time, occasionally with sparse aerial mycelium; cream-colored, red-brown, olive-brown, dark brown, or black; margins entire or fimbriate. Hyphae transversely and longitudinally septate, hyaline and thin-walled when young, frequently becoming melanized and thick-walled with age, may develop into chlamydospores. Conidiogenous cells undifferentiated, intercalary, terminal uncommon, cylindrical, with blastic conidiogenesis occurring from one to two loci per cell. Chlamydospores dark brown, smooth to lightly rough-walled, globose to ellipsoidal, septate. Conidia often yeast-like, hyaline, aseptate, smooth-walled, ellipsoidal to lemon-shaped, variable in size, indistinct hilum, budding common, polar, bipolar and multilateral.
Zalaria alba Visagie, Z. Humphries & Seifert, sp. nov.
MycoBank MB821629
(Fig. 4)
Etymology: Latin, alba, meaning white, in reference to colony appearance after 7 d of growth.
Diagnosis: Differs from Z. obscura in the inability to grow at lowered aw. Colonies remain yellowish to orange-white until it darkens after about 3 wk. Conidia appearing more slender than Z. obscura. ITS barcode: KX579093. Alternative identification markers: 28S rDNA: KX579099, RPB2: KX579105, 18S rDNA: KX579111, BenA: KX579117.
Type: Canada: Saskatchewan: Regina, isol. ex house dust, 12 Mar. 2007, E. Whitfield & K. Mwange (DAOM 734001 – holotype; DAOMC 250847 – ex-type culture).
Description: Colony diameters (mm after 7 d (14 d at 25 °C)): MEA 5–6 (8–9); MEA 5 °C microcolonies, 10 °C microcolonies, 30 °C 2–5, 35 °C 1–2, 40 °C microcolonies; MEA-5 %-NaCl no growth, MEA-10 %-NaCl no growth, MEA-15 %-NaCl no growth, MEA-24 %-NaCl no growth; OA 6–7 (11–14), PDA 7–8 (10–11), DG18 3–4 (5–7), MY1012 no growth, MY50G no growth, SNA 3–5 (7–8).
Cultural characters: Colonies on MEA at 25 °C after 7 d yeast-like, smooth and slimy, obverse yellowish white to orange white (4A2–5A2), reverse greenish grey to pale orange (1B2–5A3), generally becoming dark within 3 wk, with dark areas sometimes present after 7 d, olive-yellow to dark brown (3D6-7F5), some aerial mycelium developing after longer incubation.
Microscopic characters: Young somatic hyphae at colony periphery mostly hyaline, smooth, thin-walled, branched, transversely septate, 1.5–5 μm diam; older hyphae towards colony centre melanized, dark brown, smooth to lightly roughened, thick-walled, branched, transversely and longi-septate, 2–6.5 μm diam, often developing into chlamydospores. Conidiogenous cells undifferentiated, intercalary, rarely terminal, mostly on hyaline hyphae, producing conidia percurrently from short lateral denticles not exceeding 2 μm long. Chlamydospores dark brown, smooth to lightly rough-walled, globose to ellipsoidal, septate to aseptate, one-septate spores sometimes constricted at septum, 5.5–10 × 3–7.5 μm (x = 8 ± 0.99 μm; 5.5 ± 0.71 μm). Conidia often yeast-like, hyaline, aseptate, smooth walled, ellipsoidal to lemon-shaped, variable in size, 2.5–10 × 1.5–5 μm (x = 5.5 ± 1.53 μm, 3 ± 0.73 μm), with an indistinct hilum, budding common, polar, bipolar and multilateral.
Notes: Growth on media with lowered aw distinguishes between the two Zalaria species. Zalaria alba does not grow on MEA-5 %-NaCl, MY10-12 or MY50G after 14 d. In contrast, Z. obscura produces at least microcolonies on these media. Colony size varies significantly after 14 d on MEA at 25 °C, with Z. alba colonies (8–9 mm) more restricted than those of Z. obscura (12–14 mm). Also, Z. obscura colonies darken much faster than those of Z. alba. Microscopically these species are very similar. In general, however, spores of Z. alba seem more slender.
Additional material examined: Canada: Saskatchewan: Regina, isol. Ex house dust, 21 Aug. 2007, E. Whitfield & K. Mwange (DAOMC 250848 – culture).
Zalaria obscura Visagie, Z. Humphries & Seifert, sp. nov.
MycoBank MB821630
(Fig. 5)
Etymology: Latin obscura, dark, in reference to colony appearance after 7 d of growth.
Diagnosis: Differs from Z. alba in the ability to grow at lowered aw. Colonies dark brown to black after about 7 d. Conidia appearing less slender than Z. alba. ITS barcode: KX579094. Alternative identification markers: 28S rDNA: KX579100, RPB2: KX579106, 18S rDNA: KX579112, BenA: KX579118
Type: Canada: Saskatchewan: Regina, isol. ex house dust, 21 Aug. 2007, E. Whitfield & K. Mwange (DAOM 734002 – holotype; DAOMC 250849 – ex-type culture).
Description: Colony diameters (mm after 7 d, (14 d) at 25°C)): MEA 25 °C 7–10 (12–14); MEA 5 °C microcolonies, 10 °C microcolonies, 30 °C 3–9, 35 °C 3–8, 40 °C 1–2 mm; MEA-5 %-NaCl 2–4 (3–5), MEA-10 %-NaCl microcolonies, MEA-15 %-NaCl no growth, MEA-24 %-NaCl no growth; OA 7–8 (15–16), PDA 7–9 (10–14), DG18 3–5 (6–8), MY1012 no growth, sometimes microcolonies after prolonged incubation, MY50G no growth (microcolonies), SNA 4–5 (8–9).
Cultural characteristics: Colonies on MEA at 25 °C after 7 d yeast-like, smooth and slimy, obverse dark brown (7F5) to black with some yellowish white to orange-white (4A2–5A2), olive-yellow (3D6), and olive (1E5) areas, surface leathery after 14 d; some aerial mycelium developing after prolonged incubation.
Microscopic characters: Young somatic hyphae at colony periphery mostly hyaline, smooth, thin-walled, branched, transversely septate, 1.5–4.5 μm diam; older hyphae towards colony centre melanized, dark brown, smooth to lightly roughened, thick-walled, branched, transversely and longi-septate, 2–11 μm diam, often developing into chlamydospores. Conidiogenous cells undifferentiated, intercalary, rarely terminal, mostly on hyaline hyphae, producing conidia percurrently from short lateral denticles not exceeding 2 μm long. Chlamydospores dark brown, smooth to lightly rough walled, globose to ellipsoidal, septate to aseptate, one-septate spores sometimes constricted at septum, 5–17 μm × 3.5–8 μm (x = 8 ± 2.11 μm; 6 ± 0.89 μm). Conidia often yeast-like, hyaline, aseptate, smooth-walled, ellipsoidal to lemon-shaped, variable in size, 2.5–10 × 1.5–5.5 μm (x = 5 ± 1.4 μm; 3.5 ± 0.77 μm), with an indistinct hilum, budding common, polar, bipolar and multilateral.
Notes: See notes under Z. alba.
Additional material examined: Canada: Saskatchewan: Regina, isol. exhouse dust, 21 Aug. 2007, E. Whitfield & K. Mwange (DAOMC 250850). – USA: California: Berkeley, isol. ex house dust, 31 Mar. 2005, A. Amend, E. Whitfield & K. Mwange (DAOMC 250851, 250852).
DISCUSSION
In this paper, we describe a novel lineage comprising six black yeast-like strains isolated from house dust collected in Canada and the USA as a new genus of Dothideales, Zalaria. Mature agar colonies become dark and leathery, but are covered with slimy masses of conidia or yeast-like cells. The conidiogenous cells are undifferentiated and usually intercalary, with blastic conidiogenesis occurring on 1–2 loci per cell, giving rise to aseptate, smooth-walled, ellipsoidal to lemon-shaped conidia, that commonly bud in a polar, bipolar or multilateral pattern; dark brown, rough-walled chlamydospores are often seen. Strains were resolved at the species level into two clades, described as Z. alba and Z. obscura, with strains of the latter growing faster and with colonies darkening within 7 d. No sexual morph is known for either species.
Because of the black yeast asexual morphology, Zalaria is difficult to distinguish from Aureobasidium and Hormonema. Conidiogenesis has been used to differentiate the latter two genera, but does not consistently provide accurate identification unless combined with growth rates and physiological characters such as carbohydrate assimilation (De Hoog & Yurlova 1994, Loncaric et al. 2009, Yurlova et al. 1996). The most reliable morphological character for distinguishing these genera is the number of loci present on conidiogenous cells. Species of Zalaria and Hormonema have only 1–2 loci per cell, whereas Aureobasidium has up to 14 (De Hoog &Yurlova 1994, Yurlova et al. 1999). Distinguishing Zalaria and Hormonema based only on conidiogenesis, however, is nearly impossible. Sexual morph characters are used to distinguish Dothidea, Pringsheimia and Sydowia (Thambugala et al. 2014) with similar asexual morphs, but no sexual morph has been observed in Zalaria.
Although the two species described here can be confidently interpreted as representing a new genus, our decision to propose a new family Zalariaceae is less satisfying. Our phylogenetic analyses (Figs 1, 2, 3) consistently resolved Zalaria strains as distinct from Aureobasidiaceae and Dothideaceae. Working within the framework and concepts adopted for the order (Thambugala et al. 2014), we could either synonymise all current families or introduce a new family; we chose the latter approach. Our proposal of a new family on primarily phylogenetic grounds, in the absence of presumably more informative characters of so-far unknown sexual morphs, perpetuates but does not add to the vagueness of phenotypic characters underlying the phylogeny. However, we hope that increased sampling, especially of sexually reproducing species across Dothideomycetes, of unsequenced but known and unknown taxa, will reveal morphological or other phenotypic characters that are predictive of phylogeny, resulting in stable family and genus concepts in Dothideales.
BLAST results with Zalaria strains (Table 2) recovered 11 sequences that we consider belong to Zalaria, but that were originally identified as Aureobasidium sp. or A. pullulans. These strains or clones originated from the USA, China, Thailand, Greece, The Netherlands, Portugal, Spain, and Antarctica. Strains placed by Zalar et al. (2008) in their group 5 are identified here as Z. obscura and originate from the Norwegian arctic region. Furthermore, a BLAST search of 454 pyrosequencing data generated during our house dust project (Amend et al. 2010), revealed 21 sequences belonging to Zalaria in dust collected from Australia, Canada, New Zealand, South Africa, and the USA. Zalaria seems to have a truly worldwide distribution, and occurs on many substrates, from wood, soil, dust, sediments, cork, and subglacial ice. Understanding its ecology will be very challenging. Zalar et al. (2008) suggested that, given the highly selective conditions of the environment, their then new group 5 might be restricted to areas like Kongsfjorden in Norway. Arctic environments possess low aw because ice formation removes most of the available water, while aw is lowered further as solute ions are expelled during the freezing process (Gunde-Cimerman et al. 2003). This lack of available water is a dominating factor in microbial life in arctic regions (Gunde-Cimerman et al. 2003) and favours the growth of xerotolerant and xerophilic fungi. Given the extreme environment of a polythermal glacier, it could be hard to imagine how an organism so specifically adapted could out-compete other life-forms in more hospitable climates. However, arctic fungal species seem to have very effective dispersal strategies over long distances (Geml 2011). Combined with Zalaria’s phenotypic plasticity, melanisation and the halotolerance of Z. obscura (similar to that in Aureobasidium), these species may be capable of widespread dispersal and also survive in or on many substrates.
Before our study, the only way to identify and communicate information on strains, clones or Zalaria OTU’s was by means of UNITE’s species hypotheses (Kõljalg et al. 2013), i.e. based on 0 % threshold, SH377734.07FU represents Zalaria alba and SH377739.07FU represents Z. obscurum. With formal names now available to these species hypotheses, communicating information about these fungi and studying the extent of their distribution, habitats and possible ecological roles will be much easier.
Acknowledgments
This research was supported by grants from the Alfred P. Sloan Foundation Program on the Microbiology of the Built Environment. We would like to acknowledge Ed Whitfield and Kalima Mwange who made all isolations from house dust samples. We also thank everybody who collected house dust used during this project.
REFERENCES
- Abdel-Hafez S, Mohawed S, El-Said A. (1989) Seasonal fluctuations of soil fungi of Wadi Qena at eastern desert of Egypt. Acta Mycologica 25: 113–125. [Google Scholar]
- Adams RI, Miletto M, Taylor JW, Bruns TD. (2013) Dispersal in microbes: fungi in indoor air are dominated by outdoor air and show dispersal limitation at short distances. The ISME Journal 7: 1262–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aimanianda V, Bayry J, Bozza S, Kniemeyer O, Perruccio K, et al. (2009) Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature 460: 1117–1121. [DOI] [PubMed] [Google Scholar]
- Akaike H. (1974) A new look at the statistical model identification. IEEE Transactions on Automatic Control 19: 716–723. [Google Scholar]
- Álvarez-Rodríguez ML, Belloch C, Villa M, Uruburu F, Larriba G, Coque J-JR. (2003) Degradation of vanillic acid and production of guaiacol by microorganisms isolated from cork samples. FEMS Microbiology Letters 220: 49–55. [DOI] [PubMed] [Google Scholar]
- Amend AS, Seifert KA, Samson RA, Bruns TD. (2010) Indoor fungal composition is geographically patterned and more diverse in temperate zones than in the tropics. Proceedings of the National Academy of Sciences, USA 107: 13748–13753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen B, Frisvad JC, Søndergaard I, Rasmussen IS, Larsen LS. (2011) Associations between fungal species and water-damaged building materials. Applied and Environmental Microbiology 77: 4180–4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenz BE. (2010) Fungi in Antarctica: a circumpolar study of biodiversity in soils and historic structures. PhD, thesis, University of Minnesota. [Google Scholar]
- Barata A, Santos SC, Malfeito-Ferreira M, Loureiro V. (2012) New insights into the ecological interaction between grape berry microorganisms and Drosophila flies during the development of sour rot. Microbial Ecology 64: 416–430. [DOI] [PubMed] [Google Scholar]
- Barr ME. (2001) Revisionary studies on the Dothioraceae. Harvard Papers in Botany 6: 25–34. [Google Scholar]
- Blakeslee AF. (1915) Lindner’s roll tube method of separation cultures. Phytopathology 5: 68–69. [Google Scholar]
- Boonmak C. (2013) Diversity of yeast in water and sediment from mangrove forest in the upper coast of the gulf of Thailand. MSc this, Kasetsart University, Thailand. [Google Scholar]
- Collado J, Platas G, Paulus B, Bills GF. (2007) High-throughput culturing of fungi from plant litter by a dilution-to-extinction technique. FEMS Microbiology Ecology 60: 521–533. [DOI] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Burgess TI, Hardy GESJ, Crane C, et al. (2016) Fungal Planet description sheets: 469-557. Persoonia 37: 218–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Hoog GS, Guarro J, Gene J, Figueras MJ. (2014) Atlas of Clinical Fungi. 4th edn. Utrecht: CBS-KNAW Fungal Biodiversity Centre. [Google Scholar]
- De Hoog GS, Hermanides-Nijhof EJ. (1977) The black yeasts and allied hyphomycetes. Studies in Mycology 15: 1–140. [Google Scholar]
- De Hoog GS, Yurlova NA. (1994) Conidiogenesis, nutritional physiology and taxonomy of Aureobasidium and Hormonema. Antonie van Leeuwenhoek 65: 41–54. [DOI] [PubMed] [Google Scholar]
- Dmitriev V, Gilichinski D, Faizutdinova R, Shershunov I, Golubev W, Duda V. (1997) Occurrence of viable yeasts in 3-million-year-old permafrost in Siberia. Mikrobiologiya 66: 655–660. [Google Scholar]
- Dodman RL, Reinke JR. (1982) A selective medium for determining the population of viable conidia of Cochliobolus sativus in soil. Australian Journal of Agricultural Research 33: 287–291. [Google Scholar]
- Flannigan B, Miller JD. (2011) Microbial growth in indoor environments. In: Microorganisms in Home and Indoor Work Environments: diversity, health impacts, investigation and control (Flannigan B, Samson RA, Miller JD, eds): 57–108. London: CRC Press. [Google Scholar]
- Garber G. (2001) An overview of fungal infections. Drugs 61 (Suppl. 1): 1–12. [DOI] [PubMed] [Google Scholar]
- Geml J. (2011) Coalescent analyses reveal contrasting patterns of intercontinental gene flow in arctic-alpine and boreal-temperate fungi. In: Biogeography of Microscopic Organisms: is everything small everywhere? (Fontaneto D, ed.): 177–190. Cambridge: Cambridge University Press. [Google Scholar]
- Gerrits van den Ende AH, De Hoog GS. (1999) Variability and molecular diagnostics of the neurotropic species. Studies in Mycology 43: 151–162. [Google Scholar]
- Glass NL, Donaldson GC. (1995) Development of premier sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 61: 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gostinčar C, Ohm RA, Kogej T, Sonjak S, Turk M, et al. (2014) Genome sequencing of four Aureobasidium pullulans varieties: biotechnological potential, stress tolerance, and description of new species. BMC Genomics 15: 549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunde-Cimerman N, Sonjak S, Zalar P, Frisvad J, Diderichsen B, Plemenitaš A. (2003) Extremophilic fungi in arctic ice: a relationship between adaptation to low temperature and water activity. Physics and Chemistry of the Earth 28: 1273–1278. [Google Scholar]
- Gunde-Cimerman N, Zalar P, De Hoog S, Plemenitaš A. (2000) Hypersaline waters in salterns - natural ecological niches for halophilic black yeasts. FEMS Microbiology Ecology 32: 235–240. [DOI] [PubMed] [Google Scholar]
- Guterman H, Shabtai Y. (1996) A self-tuning vision system for monitoring biotechnological processes: I. Application to production of pullulan by Aureobasidium pullulans. Biotechnology and Bioengineering 51: 501–510. [DOI] [PubMed] [Google Scholar]
- Hermanides-Nijhof EJ. (1977) Aureobasidium and allied genera. Studies in Mycology 15: 141–177. [Google Scholar]
- Hirooka Y, Tanney JB, Nguyen HD, Seifert KA. (2016) Xerotolerant fungi in house dust: taxonomy of Spiromastix, Pseudospiromastix and Sigleria gen. nov. in Spiromastigaceae (Onygenales, Eurotiomycetes). Mycologia 108: 135–156. [DOI] [PubMed] [Google Scholar]
- Hocking AD, Pitt JI. (1980) Dichloran-glycerol medium for enumeration of xerophilic fungi from low-moisture foods. Applied and Environmental Microbiology 39: 488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höppe P, Martinac I. (1998) Indoor climate and air quality. Review of current and future topics in the field of ISB study group 10. International Journal of Biometeorology 42: 1–7. [DOI] [PubMed] [Google Scholar]
- Hyde KD, Jones EBG, Liu J-K, Ariyawansa Hiran, et al. (2013) Families of Dothideomycetes. Fungal Diversity 63: 1–313. [Google Scholar]
- Jancic S, Nguyen HD, Frisvad JC, Zalar P, Schroers HJ, et al. (2015) A taxonomic revision of the Wallemia sebi species complex. PLoS One 10: e0125933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D. (1998) Biodiversity and ecology of acidophilic microorganisms. FEMS Microbiology Ecology 27: 307–317. [Google Scholar]
- Jusino MA. (2014) The fungal communities associated with Red-cockaded Woodpeckers and their excavations: descriptive and experimental evidence of symbiosis. PhD thesis, Virginia Polytechnic Institute and State University. [Google Scholar]
- Karvala K, Toskala E, Luukkonen R, Uitti J, Lappalainen S, Nordman H. (2011) Prolonged exposure to damp and moldy workplaces and new-onset asthma. International Archives of Occupational and Environmental Health 84: 713–721. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, et al. (2013) Towards a unified paradigm for sequence-based identification of fungi. Molecular Ecology Resources 22: 5271–5277. [DOI] [PubMed] [Google Scholar]
- Kornerup A, Wanscher JH. (1967) Methuen Handbook of Colour. London: Methuen. [Google Scholar]
- Liu YJ, Whelen S, Hall BD. (1999) Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Molecular Biology and Evolution 16: 1799–1808. [DOI] [PubMed] [Google Scholar]
- Loncaric I, Oberlerchner JT, Heiss B. (2009) Phenotypic and genotypic diversity among strains of Aureobasidium pullulans in comparison with related species. Antonie van Leeuwenhoek 95: 165–178. [DOI] [PubMed] [Google Scholar]
- Lopez-Garcia P, Rodriguez-Valera F, Pedros-Alio C, Moreira D. (2001) Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature 409: 603–606. [DOI] [PubMed] [Google Scholar]
- Masclaux F, Guého E, De Hoog GS, Christen R. (1995) Phylogenetic relationships of human-pathogenic Cladosporium (Xylohypha) species inferred from partial LS rRNA sequences. Journal of Medical and Veterinary Mycology 33: 327–338. [DOI] [PubMed] [Google Scholar]
- Nguyen HD, Jancic S, Meijer M, Tanney JB, Zalar P, et al. (2015) Application of the phylogenetic species concept to Wallemia sebi from house dust and indoor air revealed by multi-locus genealogical concordance. PLoS One 10: e0120894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonneman MW, Coronado G, Thempson B, Griffith WC, Hanson JD, et al. (2012) Utilizing pyrosequencing and quantitative PCR to characterize fungal populations among house dust samples. Journal of Environmental Monitoring 14: 2038–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander AJJ. (2004) MrModeltest v2. [Program distributed by the author.] Uppsala: Evolutionary Biology Centre, Uppsala University. [Google Scholar]
- O’Donnell K, Cigelnik E. (1997) Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7: 103–116. [DOI] [PubMed] [Google Scholar]
- Piecková E, Jesenská Z. (1999) Microscopic fungi in dwellings and their health implications in humans. Annals of Agricultural and Environmental Medicine 6: 1–11. [PubMed] [Google Scholar]
- Pitt JI, Hocking AD. (2009) Fungi and Food Spoilage. 3rd edn Dordrecht: Springer. [Google Scholar]
- Rittenour WR, Ciaccio CE, Barnes CS, Kashon ML, Lemons AR, et al. (2014) Internal transcribed spacer rRNA gene sequencing analysis of fungal diversity in Kansas City indoor environments. Environmental Science Processes & Impacts 16: 33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, et al. (2012) Mrbayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RA, Houbraken J, Thrane U, Frisvad JC, Andersen B. (2010) Food and Indoor Fungi. [CBS Laboratory Manual no. 2.] Utrecht: CBS-KNAW Fungal Biodiversity Institute. [Google Scholar]
- Schoch CL, Crous PW, Groenewald JZ, Boehm EW, Burgess TI, et al. (2009) A class-wide phylogenetic assessment of Dothideomycetes. Studies in Mycology 64: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklenár F, Jurjević Ž, Zalar P, Frisvad JC, Visagie CM, et al. (2017) Phylogeny of xerophilic aspergilli (subgenus Aspergillus) and taxonomic revision of section Restricti. Studies in Mycology (in press, https://doi.org/10.1016/j.simyco.2017.09.002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slepecky RA, Starmer WT. (2009) Phenotypic plasticity in fungi: a review with observations on Aureobasidium pullulans. Mycologia 101: 823–832. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanney JB, Seifert KA. (2013) Rasamsonia pulvericola sp. nov., isolated from house dust. IMA Fungus 4: 205–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanno LK, Calderon MA, Li J, Casale T, Demoly P, Joint Allergy A. (2016) Updating Allergy and/or Hypersensitivity Diagnostic Procedures in the WHO ICD-11 Revision. Journal of Allergy and Clinical Immunology: In Practice 4: 650–657. [DOI] [PubMed] [Google Scholar]
- Thambugala KM, Ariyawansa HA, Li Y-M, Boonmee S, Hongsanan S, et al. (2014) Dothideales. Fungal Diversity 68: 105–158. [Google Scholar]
- Turchetti B, Goretti M, Branda E, Diolaiuti G, D’Agata C, et al. (2013) Influence of abiotic variables on culturable yeast diversity in two distinct Alpine glaciers. FEMS Microbiology Ecology 86: 327–340. [DOI] [PubMed] [Google Scholar]
- Van Nieuwenhuijzen EJ, Houbraken JA, Meijer M, Adan OC, Samson RA. (2016) Aureobasidium melanogenum: a native of dark biofinishes on oil treated wood. Antonie van Leeuwenhoek 109: 661–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Hirooka Y, Tanney JB, Whitfield E, Mwange K, et al. (2014) Aspergillus, Penicillium and Talaromyces isolated from house dust samples collected around the world. Studies in Mycology 78: 63–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Yilmaz N, Renaud JB, Sumarah MW, Hubka V, et al. (2017) A survey of xerophilic Aspergillus from indoor environment, including descriptions of two new section Aspergillus species producing eurotium-like sexual states. MycoKeys 19: 1–30. [Google Scholar]
- Von Arx JA, Müller E. (1975) A re-evaluation of the bitunicate ascomycetes with keys to families and genera. Studies in Mycology 9: 1–159. [Google Scholar]
- Wang CJ, Zabel RA. (1990) Identification Manual for Fungi from Utility Poles in the Eastern United States. Rockville, MD: American Type Culture Collection. [Google Scholar]
- Webb JS, Nixon M, Eastwood IM, Greenhalgh M, Robson GD, Handley PS. (2000) Fungal colonization and biodeterioration of plasticized polyvinyl chloride. Applied and Environmental Microbiology 66: 3194–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR Protocols: a guide to methods and applications (Innis MA, Gelfand DH, Sninsky JS, White TJ, eds): 315–322. San Diego: Academic Press. [Google Scholar]
- Yurlova NA, De Hoog GS, Gerrits van den AH. (1999) Taxonomy of Aureobasidium and allied genera. Studies in Mycology 43: 63–69. [Google Scholar]
- Yurlova NA, Mokrousov IV, De Hoog GS. (1995) Intraspecific variability and exopolysaccharide production in Aureobasidium pullulans. Antonie van Leeuwenhoek 68: 57–63. [DOI] [PubMed] [Google Scholar]
- Yurlova NA, Uijthof JMJ, De Hoog GS. (1996) Distinction of species in Aureobasidium and related genera by PCR-ribotyping. Antonie van Leeuwenhoek 69: 323–329. [DOI] [PubMed] [Google Scholar]
- Zalar P, Gostinčar C, De Hoog G, Uršič V, Sudhadham M, Gunde-Cimerman N. (2008) Redefinition of Aureobasidium pullulans and its varieties. Studies in Mycology 61: 21–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalar P, Novak M, De Hoog GS, Gunde-Cimerman N. (2011) Dishwashers – A man-made ecological niche accommodating human opportunistic fungal pathogens. Fungal Biology 115: 997–1007. [DOI] [PubMed] [Google Scholar]


