Abstract
A comprehensive phylogeny representing 142 described and 43 provisionally named Phytophthora species is reported here for this rapidly expanding genus. This phylogeny features signature sequences of 114 ex-types and numerous authentic isolates that were designated as representative isolates by the originators of the respective species. Multiple new subclades were assigned in clades 2, 6, 7, and 9. A single species P. lilii was placed basal to clades 1 to 5, and 7. Phytophthora stricta was placed basal to other clade 8 species, P. asparagi to clade 6 and P. intercalaris to clade 10. On the basis of this phylogeny and ancestral state reconstructions, new hypotheses were proposed for the evolutionary history of sporangial papillation of Phytophthora species. Non-papillate ancestral Phytophthora species were inferred to evolve through separate evolutionary paths to either papillate or semi-papillate species.
Keywords: oomycetes, systematics, taxonomy, evolution, plant pathology
INTRODUCTION
The genus Phytophthora has had profound impacts on human history by causing agriculturally and ecologically important plant diseases (Erwin & Ribeiro 1996). Among the most notorious Phytophthora species is P. infestans, cause of the late blight disease, which was the primary cause of the Irish potato famine from 1845 to 1852 in which approximately one million people died and 1.5 million emigrated from Ireland (Turner 2005). Another example is the sudden oak death pathogen, P. ramorum, that has killed millions of coast live oak, tanoak and Japanese larch trees, and has permanently altered the forest ecosystems in California and Oregon, USA (Goheen et al. 2002, Rizzo et al. 2002, Rizzo et al. 2005). Other species, such as P. cinnamomi, P. nicotianae, and P. sojae, can also cause highly destructive plant diseases (Erwin & Ribeiro 1996). The impact caused by Phytophthora species has continued to increase with the emergence of new pathogens and diseases. The number of species known in the genus has doubled during the past decade due to extensive surveys in previously unexplored ecosystems such as natural forests (Jung et al. 2011, 2017, Rea et al. 2010, Reeser et al. 2013, Vettraino et al. 2011), streams (Bezuidenhout et al. 2010, Brazee et al. 2017, Reeser et al. 2007, Yang et al. 2016), riparian ecosystems (Brasier et al. 2003a, 2004, Hansen et al. 2012), and irrigation systems (Hong et al. 2010, 2012, Yang et al. 2014a, b). The total number of formally named species in the genus was about 58 in 1996 (Erwin & Ribeiro 1996), but now is more than 150. In addition, some provisionally or informally named species are also expected to be formally described in the near future.
A sound taxonomic system is foundational for correctly identifying Phytophthora species and safeguarding agriculture, forestry, and natural ecosystems. Traditionally, taxonomy of the genus was based on morphological characters. A fundamental morphology-based classification of Phytophthora species was established by Waterhouse (1963) who classified the species into six groups based on the morphology of sporangia, homothallism, and configuration of antheridia. However, plasticity in morphological characters amongst isolates of individual species is significant, so is homology or homoplasy among different species. For example, isolates of P. constricta (Rea et al. 2011), P. gibbosa (Jung et al. 2011), P. lateralis (Kroon et al. 2012), P. mississippiae (Yang et al. 2013), and P. multivesiculata (Ilieva et al. 1998) all produce a mixture of semi-papillate and non-papillate sporangia. Many non-papillate species recovered from irrigation water such as Phytophthora hydropathica (Hong et al. 2010) and P. irrigata (Hong et al. 2008) were morphologically inseparable from P. drechsleri, while sequence analyses demonstrated that they are distinct species. Also, production of many morphological structures and physiological features needs specific environmental conditions, while observation of these features requires substantial training and expertise. Difficulty in obtaining important morphological data can impair accurate species identification.
With the advent of DNA sequencing, the taxonomic concept for the genus has evolved from morphology to molecular phylogeny-based (Blair et al. 2008, Cooke et al. 2000, Kroon et al. 2004, Lara & Belbahri 2011, Martin et al. 2014, Martin & Tooley 2003, Robideau et al. 2011, Villa et al. 2006). In particular, the availability of whole genome sequences from P. sojae, P. ramorum (Tyler et al. 2006) and P. infestans (Haas et al. 2009) enabled the identification of genetic markers useful for multi-locus phylogenies (Blair et al. 2008).
Cooke et al. (2000) developed the first molecular phylogeny for the genus by analyzing sequences of the internal transcribed spacer region (ITS) of 51 species. Kroon et al. (2004) constructed a phylogeny based on sequences of four nuclear and mitochondrial genes of 48 species, and Blair et al. (2008) produced a sophisticated phylogeny based on sequences of seven nuclear genetic markers. That multi-locus phylogeny divided 82 Phytophthora species into 10 phylogenetically well-supported clades. Martin et al. (2014) analyzed sequences of seven nuclear and four mitochondrial genes of 90 formally named and 17 provisional species and provided phylogenies including 10 clades, almost identical to that of Blair et al. (2008), except that P. quercina and P. sp. ohioensis were excluded from clade 4 and grouped into a potentially new clade.
A comprehensive molecular phylogeny is required to understanding the evolution of Phytophthora species. Although discordance has been found between the molecular phylogeny and the morphology-based taxonomy (Cooke et al. 2000, Ersek & Ribeiro 2010), correlations have been observed between molecular phylogenies and individual morphological and physiological traits. Recent studies indicated that species in individual clades or subclades are mostly identical in sporangial papillation, and optimum and maximum growth temperatures (Cooke et al. 2000, Kroon et al. 2012, Martin et al. 2012, Yang 2014). However, there was limited to no correlation between phylogeny and the morphology of sexual organs, such as antheridial configuration (Cooke et al. 2000, Kroon et al. 2012, Martin et al. 2012, Yang 2014). These studies have implied that divergence in sporangial morphology and variation in environmental specialization may be the keys in the evolutionary history of Phytophthora species. Nevertheless, these hypotheses need to be further tested and the exact evolutionary history of the genus Phytophthora warranted more investigation.
In this study, an expanded phylogeny, including more than 180 Phytophthora taxa, many not included in any previous phylogeny, was constructed. Sequences of seven nuclear genetic markers were used for construction of the phylogeny. In light of this phylogeny, ancestral state reconstructions were conducted on the sporangial papillation of Phytophthora species. Important evolutionary divergence events and associated changes in the sporangial morphology of Phytophthora species are discussed.
MATERIALS AND METHODS
Isolate selection
A total of 376 Phytophthora isolates representing 142 described and 43 provisionally named species, plus one isolate of each Elongisporangium undulatum (basionym: Pythium undulatum), Halophytophthora fluviatilis, and Phytopythium vexans (basionym: Pythium vexans) as outgroup taxa were included (Table 1). These included 114 ex-types (Table 2). Also included were 164 authentic isolates that were designated as representative isolates by the originators of the respective species names (Table 1). The majority of these isolates were provided by the originators of the respective species, while the rest were purchased from the Westerdijk Fungal Biodiversity Institute (CBS), Utrecht, The Netherlands.
Table 1.
Information regarding isolates used in this study. GenBank accession numbers are listed in Table S1.
|
Isolate identificationd |
Isolate origins |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (Sub)cladea | Speciesb | Papillac | CH | CBS | ATCC | IMI | WPC | MG | Typee | Host or Substrate | Location | Year | Reference |
| 1a | P. cactorum | P | 22E6 | P10194 | p25 | Rhododendron sp. | Ohio, USA | n.a.f | (Schröter 1886) | ||||
| 22E7 | 16693 | 21168 | P0715 | p6 | n.a. | UK | n.a. | ||||||
| 22E8 | 16694, MYA-3653 | 50470 | P10193 | p7 | Malus sp. | Zimbabwe | n.a. | ||||||
| P. hedraiandra | P | 33F3 | MYA-4165 | p225 | Rhododendron sp. | Minnesota, USA | 2002 | (de Cock & Lévesque 2004) | |||||
| 38C2 | Irrigation water | Virginia, USA | 2006 | ||||||||||
| 62A5 | 111725 | P19523 | T | Viburnum sp. | The Netherlands | 2001 | |||||||
| P. idaei | P | 34D4 | 971.95 | MYA-4065 | 313728 | P6767 | p220 | T | Rubus idaeus | Scotland, UK | 1987 | (Kennedy & Duncan 1995) | |
| 62A1 | 968.95 | A | Rubus idaeus | Scotland, UK | 1985 | ||||||||
| P. pseudotsugae | P | 52938 | 331662 | P10339 | T | Psendotsuga menziesii | Oregon, USA | n.a. | (Hamm & Hansen 1983) | ||||
| P. aff. hedraiandra | P | 33F4 | p226 | Rhododendron sp. | Minnesota, USA | 2003 | n.a. | ||||||
| P. aff. pseudotsugae | P | 29B3 | p185 | A | Psendotsuga menziesii | Oregon, USA | 1975 | n.a. | |||||
| 1b | P. clandestina | P | 32G1 | 347.86 | 58713, 60438 | 278933 | P3943 | p200 | T | Trifolium subterraneum | Australia | 1985 | (Taylor et al. 1985) |
| 33D8 | MYA-4064 | 287317 | p215 | A | Trifolium subterranea | Australia | 1985 | ||||||
| 38D4 | p304 | n.a. | Australia | n.a. | |||||||||
| P. iranica | P | 61J4 | 374.72 | 60237 | 158964 | P3882 | p218 | T | Solanum melongena | Iran | 1969 | (Ershad 1971) | |
| P. tentaculata | P | 29F2 | 552.96 | P8497 | A | Chrysanthemum leucanthemum | Germany | n.a. | (Kröber & Marwitz 1993) | ||||
| 30D5 | Bacopa sp. | The Netherlands | 2004 | ||||||||||
| 30G8 | MYA-3655 | Argyranthemum frutescens | Germany | 2004 | |||||||||
| 1c | P. andina | SP | 60A2 | p460 | A | Solanum betaceum | Ecuador | n.a. | (Oliva et al. 2010) | ||||
| 60A3 | p461 | A | Solanum betaceum | Ecuador | n.a. | ||||||||
| P13365 | T | Solanum brevifolium | Ecuador | 2001 | |||||||||
| P. infestans | SP | 27A8 | Solanum tuberosum | Mexico | 1992 | (De Bary 1876) | |||||||
| P10650 | Solanum tuberosum | Mexico | n.a. | ||||||||||
| P. ipomoeae | SP | 31B4 | P10226 | A | Ipomoea longipedunculata | Mexico | n.a. | (Flier et al. 2002) | |||||
| 31B5 | 109229 | P10225 | T | Ipomoea longipedunculata | Mexico | 1999 | |||||||
| 31B6 | P10227 | A | Ipomoea longipedunculata | Mexico | n.a. | ||||||||
| P. mirabilis | SP | 30C1 | 64069, MYA-4062 | P3006 | p145 | A | Mirabilis jalapa | Mexico | n.a. | (Galindo-A & Hohl 1985) | |||
| 30C2 | 64070, MYA-4063 | P3007 | p153 | A | Mirabilis jalapa | Mexico | n.a. | ||||||
| P. phaseoli | SP | 23B4 | p106 | Phaseolus lunatus | Delaware, USA | 2000 | (Thaxter 1889) | ||||||
| 35B6 | Phaseolus sp. | Delaware, USA | 2000 | ||||||||||
| P10145 | Phaseolus lunatus | Delaware, USA | n.a. | ||||||||||
| P10150 | Phaseolus lunatus | Delaware, USA | n.a. | ||||||||||
| 1 | P. nicotianae | P | 22F9 | 15410, MYA-4037 | p23 | Nicotiana tabacum | North Carolina, USA | n.a. | (Breda de Haan 1896) | ||||
| 22G1 | 15409, MYA-4036 | p22 | Nicotiana tabacum | North Carolina, USA | n.a. | ||||||||
| P10116 | Metrosideros excelsa | California, USA | 2002 | ||||||||||
| P1452 | Citrus sp. | California, USA | n.a. | ||||||||||
| 2a | P. botryosa | P | 22H8 | MYA-4059 | p44 | Heavae sp. | Thailand | n.a. | (Chee 1969) | ||||
| 46C2 | 26481 | p384 | A | Hevea brasiliensis | Thailand | n.a. | |||||||
| 62C6 | 581.69 | 136915 | P3425 | T | Hevea brasiliensis | Malaysia | 1966 | ||||||
| 130422 | P6945 | Hevea brasiliensis | Malaysia | 1986 | |||||||||
| P. citrophthora | P | 03E5 | p132 | Irrigation water | Virginia, USA | 2000 | (Smith & Smith 1906) | ||||||
| 26H3 | p31 | n.a. | n.a. | n.a. | |||||||||
| P. colocasiae | SP | 22F8 | MYA-4159 | p47 | Colocasia esculenta | n.a. | 1992 | (Raciborski 1900) | |||||
| 35D3 | p276 | Colocasia esculenta | Hawaii, USA | 2005 | |||||||||
| P. himalsilva | P | 61G2 | 128767 | T | Quercus leucotricophora | Nepal | 2005 | (Vettraino et al. 2011) | |||||
| 61G3 | 128753 | A | Quercus leucotricophora | Nepal | 2005 | ||||||||
| P. meadii | P | 22G5 | MYA-4043 | p75 | Citrus sp. | India | 1992 | (McRae 1918) | |||||
| 61J9 | 219.88 | 129185 | Hevea brasiliensis | India | 1987 | ||||||||
| P. occultans | SP | 65B9 | 101557 | T | Buxus sempervirens | The Netherlands | 1998 | (Man In’t Veld et al. 2015) | |||||
| P. terminalis | SP | 65B8 | 133865 | T | Pachysandra terminalis | The Netherlands | 2010 | (Man In’t Veld et al. 2015) | |||||
| P. aff. citrophthora | P | 26H4 | p32 | A | n.a. | n.a. | n.a. | n.a. | |||||
| 342898 | P10341 | A | Syringa sp. | England, UK | 1990 | ||||||||
| P. aff. himalsilva | P | 61G4 | 128754 | A | Castanopsis sp. | Nepal | 2005 | n.a. | |||||
| P. sp. 46C3 | n.a. | 46C3 | 66767 | P6713 | p385 | A | Hevea brasiliensis | Malaysia | n.a. | n.a. | |||
| P. sp. P6262 | n.a. | P6262 | A | Hevea brasiliensis | India | n.a. | n.a. | ||||||
| P. sp. P6310 | n.a. | P6310 | A | Theobroma cacao | Indonesia | n.a. | n.a. | ||||||
| 2b | P. capsici | P | 22F4 | 15399, MYA-4034 | p8 | A | Capsicum annum | New Mexico, USA | 1948 | (Leonian 1922) | |||
| 46012 | P0253 | Theobroma cacao | Mexico | 1964 | |||||||||
| 121656 | P10386 | Cucumis sativus | Michigan, USA | 1997 | |||||||||
| P. glovera | SP | 31E5 | p167 | A | Nicotiana tabacum | Brazil | n.a. | (Abad et al. 2011) | |||||
| 62B4 | 121969 | P11685 | T | Nicotiana tabacum | Brazil | 1995 | |||||||
| P. mengei | SP | 42B2 | MYA-4554 | p340 | T | Persea americana | California, USA | n.a. | (Hong et al. 2009) | ||||
| 42B3 | MYA-4555 | p341 | A | Persea americana | California, USA | n.a. | |||||||
| P. mexicana | P | 45G4 | 554.88 | 46731 | 92550 | P0646 | p355 | Solanum lycopersicum | Argentina | n.a. | (Hotson & Hartge 1923) | ||
| P. siskiyouensis | SP | 41B7 | 122779 | MYA-4187 | P15122 | T | Stream water | Oregon, USA | 2003 | (Reeser et al. 2007) | |||
| 41B8 | A | Soil | Oregon, USA | 2003 | |||||||||
| P. tropicalis | P | 22H5 | p27 | Vanila sp. | Tahiti | n.a. | (Aragaki & Uchida 2001) | ||||||
| 35C8 | 434.91 | 76651, MYA-4218 | p272 | T | Macadamia integrifolia | Hawaii, USA | n.a. | ||||||
| P. aff. capsici | P | 22F5 | 15427, MYA-4035 | p9 | Nicotiana tabacum | North Carolina, USA | n.a. | n.a. | |||||
| P. sp. brasiliensis | n.a. | 46705 | P0630 | A | Theobroma cacao | Brazil | 1969 | (Oudemans & Coffey 1991) | |||||
| 2c | P. acerina | SP | 61H1 | 133931 | T | Acer pseudoplatanus | Italy | 2010 | (Ginetti et al. 2014) | ||||
| 61H2 | A | Soil | Italy | 2010 | |||||||||
| P. capensis | SP | 62C1 | 128319 | P1819 | T | Curtisia dentata | South Africa | n.a. | (Bezuidenhout et al. 2010) | ||||
| 62C2 | 128320 | P1822 | A | Stream water | South Africa | n.a. | |||||||
| 62C3 | 128321 | P1823 | A | Olea campensis | South Africa | 1986 | |||||||
| P. citricola | SP | 33H8 | 221.88 | 60440 | 21173 | P0716 | p396 | T | Citrus sinensis | Taiwan | 1987 | (Sawada 1927) | |
| 33J2 | 295.29 | p375 | A | Citrus sp. | Japan | 1929 | |||||||
| P. multivora | SP | 55C5 | 124094 | T | Soil | Western Australia, Australia | 2007 | (Scott et al. 2009) | |||||
| P. pachypleura | SP | 61H6 | A | Soil | UK | 2006 | (Henricot et al. 2014) | ||||||
| 61H7 | 502404 | T | Acuba japonica | UK | 2008 | ||||||||
| 61H8 | A | Soil | UK | 2009 | |||||||||
| P. pini | SP | 22F1 | MYA-3656 | p53 | A | Rhododendron sp. | West Virginia, USA | 1987 | (Hong et al. 2011) | ||||
| 45F1 | 64532 | p343 | T | Pinus resinosa | Minnesota, USA | 1925 | |||||||
| P. plurivora | SP | 22E9 | MYA-3657 | p101 | Kalmia latifolia | Western Australia, Australia | 1998 | (Jung & Burgess 2009) | |||||
| 22F2 | p52 | Rhododendron sp. cv. “Olga Mezitt” | New York, USA | n.a. | |||||||||
| 33H9 | 379.61 | Rhododendron sp. | Germany | 1958 | |||||||||
| P. sp. 22F3 | SP | 22F3 | p33 | A | n.a. | Ohio, USA | n.a. | n.a. | |||||
| P. sp. 28D1 | SP | 28D1 | p119 | A | Fagus sylvatica | New York, USA | n.a. | n.a. | |||||
| 28D3 | p121 | A | Fagus sylvatica | New York, USA | n.a. | ||||||||
| P. sp. citricola VIII | SP | 27D9 | A | Unidentified leaf | Hainan, China | n.a. | n.a. | ||||||
| P. sp. pini-like | SP | 56G1 | A | Taxus sp. | Pennsylvania, USA | 2011 | n.a. | ||||||
| P. taxon emzansi | SP | 61F2 | A | Agathosma betulina | South Africa | 2005 | (Bezuidenhout et al. 2010) | ||||||
| 61F3 | A | Agathosma betulina | South Africa | 2005 | |||||||||
| 2d | P. bisheria | SP | 29D2 | Rubus idaeus cv. Canby | Wisconsin, USA | 1989 | (Abad et al. 2008) | ||||||
| 31E6 | 122081 | P10117 | T | Fragaria ×ananassa | North Carolina, USA | 1999 | |||||||
| P1620 | Rhododendron sp. | North Carolina, USA | n.a. | ||||||||||
| P. elongata | SP | 33J3 | A | n.a. | Australia | 1995 | (Rea et al. 2010) | ||||||
| 33J4 | A | n.a. | Australia | 1995 | |||||||||
| 55C4 | 125799 | T | Soil | Western Australia, Australia | 2004 | ||||||||
| P. frigida | P | 47G6 | A | Eucalyptus smithi | South Africa | n.a. | (Maseko et al. 2007) | ||||||
| 47G7 | A | Eucalyptus smithi | South Africa | n.a. | |||||||||
| 47G8 | T | Eucalyptus smithi | South Africa | 2001 | |||||||||
| 2e | P. multivesiculata | SP to NP | 29E3 | 545.96 | P10410 | T | Cymbidium sp. | The Netherlands | n.a. | (Ilieva et al. 1998) | |||
| 30D4 | A | Cymbidium sp. | The Netherlands | n.a. | |||||||||
| P. taxon aquatilis | SP | 38J5 | MYA-4577 | A | Stream water | Virginia, USA | 2006 | (Hong et al. 2012) | |||||
| 3 | P. ilicis | SP | 23A7 | 56615, MYA-3897 | P3939 | p113 | Ilex sp. | Canada | n.a. | (Buddenhagen & Young 1957) | |||
| 34D6 | Quercus sp. | Germany | 1999 | ||||||||||
| 62A7 | 114348 | T | Ilex aquifolium | The Netherlands | n.a. | ||||||||
| P. nemorosa | SP | 28J3 | MYA-4061 | p141 | Umbellularia californica | California, USA | n.a. | (Hansen et al. 2003) | |||||
| 41C4 | MYA-2948 | p320 | T | Lithocarpus densiflorus | California, USA | n.a. | |||||||
| P. pluvialis | SP | 60B3 | MYA-4930 | T | Rainwater | Oregon, USA | 2008 | (Reeser et al. 2013) | |||||
| P. pseudosyringae | SP | 30A8 | 111772 | MYA-4222 | p284 | T | Quercus robur | Germany | 1997 | (Jung et al. 2003) | |||
| 30B1 | Pp285 | A | Quercus robur | Germany | 1997 | ||||||||
| P. psychrophila | SP | 29J5 | 803.95 | T | Quercus robur | Germany | 1995 | (Jung et al. 2002) | |||||
| 29J6 | MYA-4083 | p288 | A | Quercus ilex | France | 1996 | |||||||
| 4 | P. alticola | P | 47G5 | 121939 | P16948 | A | Eucalyptus dunnii | South Africa | n.a. | (Maseko et al. 2007) | |||
| P. arenaria | P | 55C2 | 127950 | T | Soil | Western Australia, Australia | 2009 | (Rea et al. 2011) | |||||
| 62B7 | 125800 | A | Soil | Western Australia, Australia | 2009 | ||||||||
| P. megakarya | P | 22H7 | MYA-4040 | p42 | Theobroma cacao | Africa | n.a. | (Brasier & Griffin 1979) | |||||
| 61J5 | 238.83 | 42100 | 202077 | T | Theobroma cacao | Cameroon | n.a. | ||||||
| 61J6 | 239.83 | 42099 | 106327 | A | Theobroma cacao | Nigeria | n.a. | ||||||
| P. palmivora | P | 22G8 | MYA-4039 | P10213 | p65 | Citrus sp. | Florida, USA | n.a. | (Butler 1910) | ||||
| 22G9 | MYA-4038 | p26 | Theobroma cacao | Costa Rica | n.a. | ||||||||
| P. quercetorum | P | 15C7 | Soil | South Carolina, USA | 1997 | (Balci et al. 2008) | |||||||
| 15C8 | Soil | South Carolina, USA | 1997 | ||||||||||
| P. quercina | P | 30A4 | 783.95 | A | Quercus robur | Germany | 1995 | (Jung et al. 1999) | |||||
| 30A5 | 784.95 | MYA-4084 | T | Quercus robur | Germany | 1995 | |||||||
| 30A7 | Quercus sp. | Serbia | 2003 | ||||||||||
| P. sp. ohioensis | n.a. | P16050 | A | Soil | Ohio, USA | 2006 | n.a. | ||||||
| 5 | P. agathidicida | P | 67D5 | T | Agathis australis | New Zealand | 2006 | (Weir et al. 2015) | |||||
| P. castaneae | P | 22H6 | MYA-4060 | p45 | Castanea sp. | Japan | n.a. | (Katsura 1976) | |||||
| 30E7 | Soil | Hainan, China | n.a. | ||||||||||
| 61J7 | 587.85 | 36818 | 325914 | T | Soil | Taiwan | n.a. | ||||||
| P. cocois | P | 67D6 | T | Cocos nucifera | Hawaii, USA | 1990 | (Weir et al. 2015) | ||||||
| P. heveae | P | 22J1 | 180616 | p28 | T | Heavae sp. | Malaysia | n.a. | (Thompson 1929) | ||||
| 22J2 | 16701, MYA-3895 | p17 | soil | Tennessee, USA | 1964 | ||||||||
| 6a | P. gemini | NP | 46H1 | 123382 | A | Zostera marina | The Netherlands | 1999 | (Man in’t Veld et al. 2011) | ||||
| 46H2 | 123383 | A | Zostera marina | The Netherlands | 1999 | ||||||||
| P. humicola | NP | 32F8 | 200.81 | 52179, MYA-4080 | P3826 | p198 | T | Soil | Taiwan | 1976 | (Ko & Ann 1985) | ||
| 32F9 | P6702 | p199 | A | Phaseolus vulgaris | Taiwan | n.a. | |||||||
| P. inundata | NP | 30J3 | 390121 | p291 | T | Olea sp. | Spain | 1996 | (Brasier et al. 2003b) | ||||
| 30J4 | 389751 | p298 | T | Salix matsudana | UK | 1972 | |||||||
| P8619 | Pistacia vera | Iran | n.a. | ||||||||||
| P. rosacearum | NP | 22J9 | MYA-3662 | p82 | A | Prunus sp. | California, USA | 1987 | (Hansen et al. 2009) | ||||
| 41C1 | p321 | A | Prunus sp. | California, USA | n.a. | ||||||||
| 47J1 | MYA-4456 | T | Malus domestica | California, USA | n.a. | ||||||||
| P. sp. 48H2 | NP | 48H2 | A | Stream water | Virginia, USA | 2008 | n.a. | ||||||
| P. sp. 62C9 | NP | 62C9 | A | Stream water | Taiwan | 2013 | n.a. | ||||||
| P. sp. personii | n.a. | P11555 | A | Nicotiana tabacum | North Carolina, USA | n.a. | n.a. | ||||||
| P. taxon walnut | NP | 40A7 | A | Irrigation water | Virginia, USA | 2006 | (Brasier et al. 2003a) | ||||||
| 43G1 | A | Irrigation water | Virginia, USA | 2007 | |||||||||
| 6b | P. amnicola | NP | 61G6 | 131652 | T | Stream water | Western Australia, Australia | 2009 | (Crous et al. 2012) | ||||
| 62C5 | 133867 | Pachysandra sp. | The Netherlands | n.a. | |||||||||
| P. bilorbang | NP | 61G8 | 131653 | T | Soil | Western Australia, Australia | 2010 | (Aghighi et al. 2012) | |||||
| P. borealis | NP | 60B2 | 132023 | MYA-4881 | T | Stream water | Alaska, USA | 2008 | (Hansen et al. 2012) | ||||
| P. crassamura | NP | 66C9 | A | Picea abies | Italy | 2012 | (Scanu et al. 2015) | ||||||
| 66D1 | 140357 | T | Soil | Italy | 2011 | ||||||||
| P. fluvialis | NP | 55B6 | 129424 | T | Stream water | Western Australia, Australia | 2009 | (Crous et al. 2011) | |||||
| P. gibbosa | NP to SP | 55B7 | A | Soil | Western Australia, Australia | 2009 | (Jung et al. 2011) | ||||||
| 62B8 | 127951 | T | Soil | Western Australia, Australia | 2009 | ||||||||
| P. gonapodyides | NP | 21J5 | 46726 | p117 | Water | England, UK | n.a. | (Buisman 1927, Petersen 1910) | |||||
| 34A8 | 554.67 | 60351 | P6872 | Reservoir water | n.a. | 1967 | |||||||
| P. gregata | NP | 55B8 | A | Soil | Western Australia, Australia | 2009 | (Jung et al. 2011) | ||||||
| 62B9 | 127952 | T | Soil | Western Australia, Australia | 2009 | ||||||||
| P. lacustris | NP | 61D6 | A | Soil | Germany | 2003 | (Nechwatal et al. 2013) | ||||||
| 61D8 | A | Soil | Germany | 2003 | |||||||||
| NP | 61E1 | A | Soil | Germany | 2006 | ||||||||
| 389725 | P10337 | T | Salix matsudana | England, UK | 1972 | ||||||||
| P. litoralis | NP | 55B9 | 127953 | T | Soil | Western Australia, Australia | 2008 | (Jung et al. 2011) | |||||
| P. megasperma | NP | 62C7 | 402.72 | 58817 | 32035 | P3599 | T | Althaea rosea | Washington DC, USA | 1931 | (Drechsler 1931) | ||
| P. mississippiae | NP to SP | 57J1 | A | Irrigation water | Mississippi, USA | 2012 | (Yang et al. 2013) | ||||||
| 57J2 | A | Irrigation water | Mississippi, USA | 2012 | |||||||||
| 57J3 | MYA-4946 | T | Irrigation water | Mississippi, USA | 2012 | ||||||||
| 57J4 | A | Irrigation water | Mississippi, USA | 2012 | |||||||||
| P. ornamentata | NP | 66D2 | 140647 | T | Soil | Italy | 2012 | (Scanu et al. 2015) | |||||
| 66D3 | A | Soil | Italy | 2012 | |||||||||
| P. pinifolia | NP | 47H1 | 122924 | T | Pinus radiata | Chile | 2007 | (Duran et al. 2008) | |||||
| 47H2 | 122922 | A | Pinus radiata | Chile | 2007 | ||||||||
| P. riparia | NP | 60B1 | 132024 | MYA-4882 | T | Stream water | Oregon, USA | 2006 | (Hansen et al. 2012) | ||||
| P. thermophila | NP | 55C1 | 127954 | T | Soil | Western Australia, Australia | 2004 | (Jung et al. 2011) | |||||
| P. ×stagnum | NP | 36H8 | A | Irrigation water | Virginia, USA | 2006 | (Yang et al. 2014c) | ||||||
| 36J7 | A | Irrigation water | Virginia, USA | 2006 | |||||||||
| 43F3 | MYA-4926 | T | Irrigation water | Virginia, USA | 2007 | ||||||||
| 44F9 | A | Irrigation water | Virginia, USA | 2007 | |||||||||
| P. sp. 26E1 | NP | 26E1 | p116 | A | Malus domestica | New York, USA | n.a. | n.a. | |||||
| P. sp. canalensis | n.a. | P10456 | A | Canal water | California, USA | 2002 | n.a. | ||||||
| P. sp. delaware | NP | 63H4 | A | Pond water | Delaware, USA | 2014 | n.a. | ||||||
| 63H7 | A | Pond water | Delaware, USA | 2014 | |||||||||
| P. sp. gregata-like | NP | 22J5 | 16698 | p16 | A | n.a. | n.a. | n.a. | n.a. | ||||
| P. sp. megasperma-like | NP | 23A1 | p81 | A | Prunus sp. | California, USA | n.a. | n.a. | |||||
| 23A3 | MYA-3660 | p79 | A | Actinidia chinensis | California, USA | 1987 | |||||||
| 6 | P. asparagi | NP | 33D7 | 384046 | A | Asparagus officinalis | New Zealand | 1980 | (Crous et al. 2012) | ||||
| 62C4 | 132095 | MYA-4826 | T | Asparagus officinalis | Michigan, USA | 2006 | |||||||
| P. sp. sulawesiensis | n.a. | P6306 | A | Syzygium aromaticum | Indonesia | 1989 | n.a. | ||||||
| 7a | P. attenuata | NP | 67C5 | T | Soil | Taiwan | 2013 | (Jung et al. 2017) | |||||
| P. europaea | NP | 30A3 | Quercus sp. | France | 1998 | (Jung et al. 2002) | |||||||
| 34C2 | Quercus sp. | Germany | 1999 | ||||||||||
| 62A2 | 109049 | T | Soil | France | 1998 | ||||||||
| P. flexuosa | NP | 67C3 | T | Soil | Taiwan | 2013 | (Jung et al. 2017) | ||||||
| P. formosa | NP | 67C4 | T | Soil | Taiwan | 2013 | (Jung et al. 2017) | ||||||
| P. fragariae | NP | 22G6 | 11374 | P3570 | p114 | Fragaria ×ananassa | Maryland, USA | n.a. | (Hickman 1940) | ||||
| 30C5 | Fragaria ×ananassa | Virginia, USA | n.a. | ||||||||||
| 61J3 | 209.46 | 181417 | P6231 | T | Fragaria ×ananassa | England, UK | n.a. | ||||||
| P. intricata | NP | 67B9 | T | Soil | Taiwan | 2013 | (Jung et al. 2017) | ||||||
| P. rubi | NP | 30D7 | p186 | A | Rubus sp. | Australia | n.a. | (Man in ‘t Veld 2007) | |||||
| 41D5 | Rubus sp. | Norway | 2005 | ||||||||||
| 46C7 | 90442 | p389 | T | Rubus idaeus cv. "Glen Clova" | Scotland, UK | n.a. | |||||||
| P. uliginosa | NP | 62A3 | 109054 | P10413 | T | Soil | Poland | 1998 | (Jung et al. 2002) | ||||
| 62A4 | 109055 | P10328 | A | Soil | Germany | 1998 | |||||||
| P. ×alni | NP | 32J6 | 392317 | MYA-4081 | p205 | A | Alnus glutinosa | France | 1996 | (Brasier et al. 2004, Husson et al. 2015) | |||
| 32J7 | 392318 | p206 | A | Alnus sp. | Austria | 1996 | |||||||
| 47A7 | 392314 | T | Alnus sp. | UK | 1994 | ||||||||
| 47A8 | A | Alnus sp. | The Netherlands | n.a. | |||||||||
| P. ×cambivora | NP | 22F6 | 46719, MYA-4076 | p64 | Abies sp. | Oregon, USA | n.a. | (Buisman 1927, Jung et al. 2017) | |||||
| 26F8 | MYA-4075 | p38 | n.a. | New York, USA | n.a. | ||||||||
| P. ×heterohybrida | NP | 67C1 | T | Stream water | Taiwan | 2013 | (Jung et al. 2017) | ||||||
| P. ×incrassata | NP | 67C2 | T | Stream water | Taiwan | 2013 | (Jung et al. 2017) | ||||||
| P. sp. europaea SW | NP | 33F7 | p229 | A | Soil | West Virginia, USA | 2005 | n.a. | |||||
| 7b | P. asiatica | NP | 45G1 | 90455 | p352 | A | Robinia pseudoacacia | Jiangsu, China | n.a. | (Rahman et al. 2014a) | |||
| 46C6 | 56194 | p388 | A | Robinia pseudoacacia | Jiangsu, China | n.a. | |||||||
| 61H3 | 133347 | T | Pueraria lobata | Japan | 2005 | ||||||||
| P. cajani | NP | 33D9 | p214 | Cajanus cajani | India | n.a. | (Amin et al. 1978) | ||||||
| 45F6 | 44389 | p348 | A | Cajanus cajani | India | n.a. | |||||||
| 45F7 | 44388 | P3105 | p349 | T | Cajanus cajani | India | n.a. | ||||||
| P. melonis | NP | 32F6 | MYA-4079 | P1371 | p196 | A | Cucumis sativus | China | n.a. | (Katsura 1976) | |||
| 41B4 | p318 | A | Cucumis sativus | Iran | n.a. | ||||||||
| 45F3 | 582.69 | 52854 | T | Cucumis sativus | Japan | n.a. | |||||||
| P. niederhauserii | NP | 01D5 | p312 | A | Irrigation water | Virginia, USA | 2000 | (Abad et al. 2014) | |||||
| 23J6 | MYA-4163 | p57 | A | Unknown ornamental | Israel | n.a. | |||||||
| 31E7 | P10617 | p169 | A | Thuja occidentalis | North Carolina, USA | 2001 | |||||||
| P. pisi | NP | 60A4 | T | Pea | Sweden | 2009 | (Heyman et al. 2013) | ||||||
| 60A5 | A | Pea | Sweden | 2009 | |||||||||
| P. pistaciae | NP | 33D6 | MYA-4082 | 386658 | p216 | T | Pistacia vera | Iran | 1986 | (Mirabolfathy et al. 2001) | |||
| 41A9 | p314 | A | Pistacia vera | Iran | n.a. | ||||||||
| P. sojae | NP | 22D8 | 312.62 | 16705, MYA-3899 | 131375 | p19 | Glycine max | Ontario, Canada | 1959 | (Kaufmann & Gerdemann 1958) | |||
| 28F9 | p236 | Glycine max | Mississippi, USA | 1970 | |||||||||
| P. vignae | NP | 45G6 | 46735 | p357 | A | Glycine max | n.a. | n.a. | (Purss 1957) | ||||
| 45G9 | 64832 | 316196 | P3420 | p379 | Vigna unguiculata | Sri Lanka | n.a. | ||||||
| 46C1 | 112.76 | 64129 | p380 | Vigna sinensis | n.a. | n.a. | |||||||
| 7c | P. cinnamomi | NP | 23B1 | 15400, MYA-4057 | p10 | Camellia japonica | South Carolina, USA | n.a. | (Rands 1922) | ||||
| 23B2 | 15401, MYA-4058 | p11 | Persea americana | Puerto Rico | 1960 | ||||||||
| 61J1 | 144.22 | 46671 | 22938 | P2110 | T | Cinnamomum burmannii | Indonesia | 1922 | |||||
| P. parvispora | NP | 30G9 | MYA-4078 | p178 | A | Beaucarnea sp. | Germany | 1991 | (Scanu et al. 2014) | ||||
| 46F6 | A | Beaucarnea sp. | Germany | 1992 | |||||||||
| 66C7 | 132771 | A | Arbutus unedo | Italy | 2008 | ||||||||
| 66C8 | 132772 | T | Arbutus unedo | Italy | 2011 | ||||||||
| P. sp. ax | NP | 46H5 | A | Ilex glabra cv. “Shamrock” | Virginia, USA | 2008 | n.a. | ||||||
| 7d | P. fragariaefolia | NP | 61H4 | 135747 | T | Fragaria ×ananassa | Japan | 2005 | (Rahman et al. 2014b) | ||||
| P. nagaii | NP | 61H5 | 133248 | T | Rosa sp. | Japan | 1968 | (Rahman et al. 2014b) | |||||
| 8a | P. cryptogea | NP | 61H9 | 113.19 | 180615 | P1738 | T | Solanum lycopersicum | Ireland | n.a. | (Pethybridge & Lafferty 1919) | ||
| P. drechsleri | NP | 15E5 | Soil | South Carolina, USA | 1997 | (Tucker 1931) | |||||||
| 15E6 | Soil | South Carolina, USA | 1998 | ||||||||||
| 23J5 | 292.35 | 46724 | P1087 | p41 | T | Beta vulgaris var. altissima | California, USA | n.a. | |||||
| P10331 | Gerbera jamesonii | New Hampshire, USA | 2003 | ||||||||||
| P. erythroseptica | NP | 61J2 | 129.23 | 34684 | P1693 | T | Solanum tuberosum | Ireland | n.a. | (Pethybridge 1913) | |||
| P. medicaginis | NP | 23A4 | MYA-3900 | p37 | Medicago sativa | Ohio, USA | n.a. | (Hansen & Maxwell 1991) | |||||
| 28F1 | 44390 | P1057 | p124 | Medicago sativa | California, USA | 1975 | |||||||
| P. pseudocryptogea | NP | 52402 | P3103 | Solanum marginatum | Ecuador | n.a. | (Safaiefarahani et al. 2015) | ||||||
| P. richardiae | NP | 31E8 | P10355 | p170 | Zantedeschia sp. | Japan | 1989 | (Buisman 1927) | |||||
| 45F5 | 240.30 | 60353, 46734 | 325930 | p347 | T | Zantedeschia aethiopica | USA | n.a. | |||||
| P10811 | Zantedeschia aethiopica | Japan | 1989 | ||||||||||
| P. sansomeana | NP | 47H3 | MYA-4455 | T | Glycine sp. | Indiana, USA | n.a. | (Hansen et al. 2009) | |||||
| 47H4 | A | Glycine sp. | Indiana, USA | n.a. | |||||||||
| 47H5 | A | Glycine sp. | Indiana, USA | n.a. | |||||||||
| P. trifolii | NP | 29B2 | MYA-3901 | p142 | A | Trifolium vesiculosum | Mississippi, USA | 1978 | (Hansen & Maxwell 1991) | ||||
| 62A9 | 117687 | T | Trifolium sp. | Mississippi, USA | n.a. | ||||||||
| P. aff. cryptogea | NP | 22G2 | 308.62 | 15402, MYA-4161 | 325907 | p12 | Aster sp. | California, USA | n.a. | n.a. | |||
| P. aff. erythroseptica | NP | 22J4 | MYA-4041 | p50 | n.a. | Ohio, USA | n.a. | n.a. | |||||
| 33A1 | p207 | Solanum tuberosum | Maine, USA | 2004 | |||||||||
| P. sp. kelmania | NP | 24A7 | MYA-4162 | p102 | A | Abies concolor | West Virginia, USA | 1998 | n.a. | ||||
| 31E4 | P10613 | p166 | A | Abes fraseri | North Carolina, USA | 2002 | |||||||
| 8b | P. brassicae | SP | 29D8 | 686.95 | A | Brassica oleracea | The Netherlands | 1995 | (Man in’t Veld et al. 2002) | ||||
| 61J8 | 179.87 | P7517, P19521 | T | Brassica oleracea | The Netherlands | 1986 | |||||||
| P. cichorii | SP | 62A8 | 115029 | T | Cichorium intybus var. foliosum | The Netherlands | 2004 | (Bertier et al. 2013) | |||||
| P. dauci | SP | 61E5 | 127102 | T | Daucus carota | France | 2009 | (Bertier et al. 2013) | |||||
| 32E5 | Duscus carota | France | 2004 | ||||||||||
| 32E6 | P10728 | Duscus carota | France | 2004 | |||||||||
| 32E7 | p194 | Duscus carota | France | 2004 | |||||||||
| P. lactucae | SP | 61F4 | T | Lactuca sativa | Greece | 2001 | (Bertier et al. 2013) | ||||||
| 61F7 | A | Lactuca sativa | Greece | 2002 | |||||||||
| 61F8 | A | Lactuca sativa | Greece | 2003 | |||||||||
| P. primulae | SP | 29E9 | 620.97 | p286 | Primula acaulis | Germany | 1997 | (Tomlinson 1952) | |||||
| 29F1 | p287 | Primula sp. | The Netherlands | 1998 | |||||||||
| P. aff. brassicae-2 | n.a. | 112968 | P6207 | A | Allium cepa | Switzerland | n.a. | n.a. | |||||
| P. aff. cichorii | SP | 61E3 | 133815 | A | Cichorium intybus var. foliosum | UK | 1999 | n.a. | |||||
| P. sp. 29E7 | SP | 29E7 | A | Allium porrum | The Netherlands | n.a. | n.a. | ||||||
| P. taxon castitis | SP | 61E7 | 131246 | A | Fragaria ×ananassa | Sweden | 1995 | (Bertier et al. 2013) | |||||
| P. taxon parsley | SP | 61G1 | A | Petroselinum crispum | Greece | 2006 | (Bertier et al. 2013) | ||||||
| 8c | P. foliorum | SP | 49J8 | 121655 | MYA-3638 | P10974 | T | Rhododendron sp. | Tennessee, USA | 2004 | (Donahoo et al. 2006) | ||
| P. hibernalis | SP | 22H1 | 270.31 | 60352 | 36906 | P6871 | p115 | Citrus sinensis | Portugal | 1931 | (Carne 1925) | ||
| 32F7 | 114104 | 56353, MYA-3896 | 134760 | P3822 | p197 | Citrus sinensis | Western Australia, Australia | 1958 | |||||
| P. lateralis | NP to SP | 22H9 | MYA-3898 | p51 | A | Chamaecyparis lawsoniana | Oregon, USA | n.a. | (Tucker & Milbrath 1942) | ||||
| 29A9 | 201856 | p128 | Chamaecyparis lawsoniana | California, USA | 1997 | ||||||||
| P. ramorum | SP | 32G2 | Camellia japonica | South Carolina, USA | n.a. | (Werres et al. 2001) | |||||||
| 33F2 | Quercus agrifolia | California, USA | n.a. | ||||||||||
| 8d | P. austrocedrae | SP | 41B5 | MYA-4073 | A | Austrocedrus chilensis | Argentina | n.a. | (Greslebin et al. 2007) | ||||
| 41B6 | 122911 | MYA-4074 | T | Austrocedrus chilensis | Argentina | 2005 | |||||||
| P. obscura | SP | 60E9 | 129273 | T | Soil | Germany | 1994 | (Grünwald et al. 2012) | |||||
| 60F1 | A | Pieris sp. | Oregon, USA | 2009 | |||||||||
| 60F2 | A | Kalmia latifolia | Oregon, USA | n.a. | |||||||||
| P. syringae | SP | 21H9 | 34002 | P0649 | p187 | Citrus sp. | California, USA | n.a. | (Klebahn 1905) | ||||
| 23A6 | MYA-3659 | p35 | n.a. | New York, USA | n.a. | ||||||||
| 8 | P. stricta | NP | 58A1 | MYA-4944 | T | Irrigation water | Mississippi, USA | 2012 | (Yang et al. 2014a) | ||||
| 58A2 | A | Irrigation water | Mississippi, USA | 2012 | |||||||||
| 58A3 | A | Irrigation water | Mississippi, USA | 2012 | |||||||||
| 58A4 | A | Irrigation water | Mississippi, USA | 2012 | |||||||||
| 9a (cluster 9a1) | P. aquimorbida | NP | 40A6 | MYA-4578 | T | Irrigation water | Virginia, USA | 2006 | (Hong et al. 2012) | ||||
| 40E3 | A | Irrigation water | Virginia, USA | 2006 | |||||||||
| 44G9 | A | Irrigation water | Virginia, USA | 2007 | |||||||||
| P. chrysanthemi | NP | 61E9 | A | Chrysanthemum sp. | Japan | 1998 | (Naher et al. 2011) | ||||||
| 61F1 | 123163 | T | Chrysanthemum ×morifolium | Japan | 2000 | ||||||||
| P. hydrogena | NP | 44G8 | A | Irrigation water | Virginia, USA | 2007 | (Yang et al. 2014b) | ||||||
| 46A3 | MYA-4919 | T | Irrigation water | Virginia, USA | 2007 | ||||||||
| 46A4 | A | Irrigation water | Virginia, USA | 2007 | |||||||||
| P. hydropathica | NP | 05D1 | MYA-4460 | p366 | T | Irrigation water | Virginia, USA | 2000 | (Hong et al. 2010) | ||||
| 5C11 | MYA-4459 | p365 | A | Irrigation water | Virginia, USA | 2000 | |||||||
| P. irrigata | NP | 04E4 | MYA-4458 | p335 | A | Irrigation water | Virginia, USA | 2000 | (Hong et al. 2008) | ||||
| 23J7 | MYA-4457 | p108 | T | Irrigation water | Virginia, USA | 2000 | |||||||
| 44E4 | A | Stream water | Virginia, USA | 2007 | |||||||||
| P. macilentosa | NP | 58A5 | A | Irrigation water | Mississippi, USA | 2012 | (Yang et al. 2014a) | ||||||
| 58A6 | A | Irrigation water | Mississippi, USA | 2012 | |||||||||
| 58A7 | MYA-4945 | T | Irrigation water | Mississippi, USA | 2012 | ||||||||
| 58A8 | A | Irrigation water | Mississippi, USA | 2012 | |||||||||
| P. parsiana | NP | 47C3 | 395329 | T | Ficus carica | Iran | 1991 | (Mostowfizadeh-Ghalamfarsa et al. 2008) | |||||
| P. virginiana | NP | 40A9 | A | Irrigation water | Virginia, USA | 2006 | (Yang & Hong 2013) | ||||||
| 44G6 | A | Irrigation water | Virginia, USA | 2007 | |||||||||
| 46A2 | MYA-4927 | T | Irrigation water | Virginia, USA | 2007 | ||||||||
| P. aff. parsiana G1 | NP | 47C7 | A | Pistacia vera | Iran | n.a. | n.a. | ||||||
| 47C8 | A | Pistacia vera | Iran | n.a. | |||||||||
| 395328 | P8618 | A | Pistacia vera | Iran | 1992 | ||||||||
| P. aff. parsiana G2 | NP | 47C5 | 395330 | A | Pistacia vera | Iran | 1992 | n.a. | |||||
| 47C6 | 395331 | A | Pistacia vera | Iran | 1992 | ||||||||
| P. aff. parsiana G3 | NP | 47D5 | A | Pistacia vera | Iran | n.a. | n.a. | ||||||
| 47D8 | A | Pistacia vera | Iran | n.a. | |||||||||
| 47E1 | A | Pistacia vera | Iran | n.a. | |||||||||
| P. sp. 35G4 | NP | 35G4 | A | Irrigation water | Virginia, USA | 2005 | n.a. | ||||||
| P. sp. 38D9 | NP | 38D9 | A | Dianthus caryophyllus | Taiwan | n.a. | n.a. | ||||||
| P. sp. 40J5 | NP | 40J5 | A | Unknown leaf in seawater | Hainan, China | n.a. | n.a. | ||||||
| P. sp. cuyabensis | n.a. | P8213 | A | n.a. | Ecuador | 1993 | n.a. | ||||||
| P. sp. lagoariana | NP | 60B4 | P8220 | A | n.a. | Ecuador | n.a. | n.a. | |||||
| 60B5 | P8217 | T | n.a. | Ecuador | n.a. | ||||||||
| P8223 | A | n.a. | Ecuador | 1993 | |||||||||
| 9a (cluster 9a2) | P. macrochlamydospora-G1 | SP | 33E1 | P10264 | Glycine max | New South Wales, Australia | n.a. | (Irwin 1991) | |||||
| P10267 | Glycine max | New South Wales, Australia | 1994 | ||||||||||
| P. macrochlamydospora-G2 | SP | 31E9 | 351473 | P8017 | p171 | Glycine max | Queensland, Australia | n.a. | (Irwin 1991) | ||||
| 33D5 | 240.30 | 60353 | 340618 | Zantedeschia aethiopica | The Netherlands | 1927 | |||||||
| P. quininea | NP | 45F2 | 406.48 | 56964 | p344 | A | Cinchona officinalis | Peru | n.a. | (Crandall 1947) | |||
| 46C4 | 407.48 | 46733 | p386 | T | Cinchona officinalis | Peru | n.a. | ||||||
| 9a (cluster 9a3) | P. insolita | NP | 327E1 | MYA-4077 | p123 | Waterfall water | Hainan, China | n.a. | (Ann & Ko 1980) | ||||
| 38E1 | 691.79 | 38789 | 288805 | T | Soil | Taiwan | 1980 | ||||||
| P6703 | A | Soil | Taiwan | n.a. | |||||||||
| P. polonica | NP | 40G9 | Irrigation water | Virginia, USA | 2006 | (Belbahri et al. 2006) | |||||||
| 43F9 | Irrigation water | Virginia, USA | 2007 | ||||||||||
| 49J9 | P15005 | A | Soil | Poland | 2006 | ||||||||
| 9b | P. captiosa | NP | 46H6 | A | Eucalyptus saligna | New Zealand | 1999 | (Dick et al. 2006) | |||||
| 46H7 | P10719 | T | Eucalyptus saligna | New Zealand | 1992 | ||||||||
| 46H8 | A | Eucalyptus saligna | New Zealand | 2000 | |||||||||
| P10721 | A | Eucalyptus saligna | New Zealand | 1998 | |||||||||
| P. constricta | NP to SP | 55C3 | 125801 | T | Soil | Western Australia, Australia | 2006 | (Rea et al. 2011) | |||||
| P. fallax | NP | 46J2 | P10722 | T | Eucalyptus delegatensis | New Zealand | 1997 | (Dick et al. 2006) | |||||
| 46J3 | A | Eucalyptus nitens | New Zealand | 2000 | |||||||||
| 46J5 | A | Eucalyptus nitens | New Zealand | 2000 | |||||||||
| P10725 | A | Eucalyptus fastigata | New Zealand | 2004 | |||||||||
| 10 | P. boehmeriae | P | 45F9 | 291.29 | 180614 | P6950 | T | Boehmeriae nivea | Taiwan | 1927 | (Sawada 1927) | ||
| P. gallica | NP | 50A1 | 111474 | P16826 | T | Quercus robur | France | 1998 | (Jung & Nechwatal 2008) | ||||
| 61D5 | 111475 | P16827 | A | Phragmites australis | Germany | 2004 | |||||||
| P. gondwanensis | P | 22G7 | MYA-3893 | n.a. | Ohio, USA | n.a. | (Crous et al. 2015) | ||||||
| P. intercalaris | NP | 45B7 | 140632 | TSD-7 | T | Stream water | Virginia, USA | 2007 | (Yang et al. 2016) | ||||
| 48A1 | A | Stream water | Virginia, USA | 2008 | |||||||||
| 49A7 | 140631 | A | Stream water | Virginia, USA | 2009 | ||||||||
| P. kernoviae | P | 46C8 | P10956 | p390 | Rhododendron ponticum | England, UK | 2004 | (Brasier et al. 2005) | |||||
| 46J6 | P10681 | Annona cherimola | New Zealand | 2002 | |||||||||
| 46J8 | P10671 | Soil | New Zealand | 2003 | |||||||||
| P. morindae | P | 62B5 | 121982 | T | Morinda citrifolia var. citrifolia | Hawaii, USA | 2005 | (Nelson & Abad 2010) | |||||
| P. sp. boehmeriae-like | P | 45F8 | 357.52 | 60173 | 32199 | P1378 | p350 | A | Citrus sinensis | Argentina | 1939 | n.a. | |
| n.a. | P. lilii | NP | 135746 | T | Lilium sp. | Japan | 1987 | (Rahman et al. 2015) | |||||
| outgroup | Elongisporangium undulatum | P | 101728 | 337230 | P10342 | T | Larix sp. | Scotland, UK | 1989 | (Uzuhashi et al. 2010) | |||
| Phytopythium vexans | P | 340.49 | 12194 | P3980 | T | n.a. | n.a. | n.a. | (de Cock et al. 2015) | ||||
| Halophytophthora fluviatilis | P | 57A9 | MYA-4961 | T | Stream water | Virginia, USA | 2011 | (Yang & Hong 2014) | |||||
a Molecular (sub)clade as designated in Fig. 1
b Names of taxa informally designated for the first time in this study are underlined.
c Sporangial papillation: NP = non-papillate, P = papillate, and SP = semi-papillate.
d Isolate identification abbreviations: CH, Chuanxue Hong laboratory at Virginia Polytechnic Institute and State University, Virginia Beach, VA, USA; CBS, Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands; ATCC, American Type Culture Collection, Manassas, VA, USA; IMI, CABI Biosciences, UK; WPC, the World Phytophthora Genetic Resource Collection at University of California, Riverside, USA; MG, Mannon E. Gallegly laboratory at West Virginia University, USA. Local identifications of respective isolates are provided in Table S1.
e Ex-types (T) or authentic (A) isolates (designated as representative isolates by the originators of the respective species).
f n.a.= not available.
Table 2.
Numbers of species and ex-types included in phylogenies for the genus Phytophthora in previous studies and this study.
|
Number of species |
|||
|---|---|---|---|
| Phylogeny in | Formal | Provisional | Number of ex-types |
| Cooke et al. (2000) | 49 | 2 | 9 |
| Kroon et al. (2004) | 46 | 2 | 18 |
| Blair et al. (2008) | 72 | 10 | 16 |
| Martin et al. (2014) | 90 | 17 | 31 |
| This study | 142 | 43 | 114 |
DNA extraction
To extract genomic DNA (gDNA), an approximately 5 × 5 mm culture plug of each isolate was taken from the actively growing area of a fresh culture. This was then grown in 20 % clarified V8 broth (lima bean broth for growing a P. infestans isolate 27A8) at room temperature (ca. 23 °C) for 7–14 d to produce a mycelial mass. The mass was then blot-dried using sterile tissue paper and then lysed in liquid nitrogen or using a FastPrep®-24 system (MP Biomedicals, Santa Ana, CA). gDNA was extracted using the DNeasy® Plant Mini kit (Qiagen, Valencia, CA) or the Maxwell® Plant DNA kit in combination with a Maxwell® Rapid Sample Concentrator (Promega, Madison, WI).
DNA amplification and sequencing
A set of primers for seven genetic markers were used for DNA amplification including 60S Ribosomal protein L10 (60S), beta-tubulin (Btub), elongation factor 1 alpha (EF1α), enolase (Enl), heat shock protein 90 (HSP90), 28S ribosomal DNA (28S), and tigA gene fusion protein (TigA) as indicated in Blair et al. (2008). PCR reaction mixtures were prepared with the Takara Taq DNA polymerase (Takara Shuzo, Shiga, Japan) according to the manufacturer’s instructions. The PCR cycling protocol was the same as indicated by Blair et al. (2008), except that the Eppendorf® Mastercycler® Pro thermal cycler (Eppendorf, Hamburg) was used in this study. All PCR products were evaluated for successful amplification using agarose gel electrophoresis. Unsuccessful PCR amplifications were repeated using a modified protocol to attempt successful amplifications by optimizing annealing temperature using gradient PCR (typically with lower annealing temperatures) or using the GoTaq® Flexi DNA Polymerase (Promega, Madison, WI) PCR mixture system.
Prior to sequencing, excess primer and dNTPs were removed from successful PCR products with shrimp alkaline phosphatase and exonuclease I (USB Catalog # 70092Y and 70073Z). One unit of each enzyme was added to 15 μL PCR product, incubated at 37 °C for 30 min, followed by heat inactivation at 65 °C for 15 min. Sequencing was performed with both amplifying primers as well as internal primers, if any, for individual genetic markers at the University of Kentucky Advanced Genetic Technologies Center (Lexington, KY). Derived sequencing files were visualized with FinchTV version 1.4.0 (Geospiza, Seattle, WA). Sequences of each isolate with all primers for individual genetic markers were aligned with Clustal W (Larkin et al. 2007) and edited manually to correct obvious sequencing errors and code ambiguous sites according to the International Union of Pure and Applied Chemistry (IUPAC) nucleotide ambiguity codes to produce a consensus sequence. All sequences produced in this study have been deposited in GenBank (Supplementary Table 1).
Among 379 isolates (including three isolates of the outgroup taxa) in the following phylogenetic analyses, all seven phylogenetic markers from 321 isolates were sequenced in this study. Sequences of all markers from 49 isolates by Blair et al. (2008) were also included in the analyses. Additionally, for seven isolates, sequences of one or two genes were newly produced in this study while the remaining gene sequences were from Blair et al. (2008). Sequences from P. lilii (CBS 135746) and P. sp. ohioensis (ST18-37) were obtained from Rahman et al. (2015) and from the Phytophthora Database (Park et al. 2013), respectively.
Phylogenetic analyses
Concatenated sequences of all isolates were aligned using Clustal X version 2.1 (Larkin et al. 2007). The alignment was edited in BioEdit version 7.2.5 (Hall 1999) to trim aligned concatenated sequences to an equal size and set missing data to question marks. The edited alignment was then analyzed in jModelTest version 2.1.7 (Posada 2008) to select the most appropriate model for the following phylogenetic analyses. Maximum likelihood (ML) analysis was performed using RAxML version 8.2.0 (Stamatakis 2014) with the selected model and 1000 bootstrap replicates. Maximum parsimony (MP) analysis was conducted using PAUP version 4.0a147 (Swofford 2002) with 1000 bootstrap replicates. Bayesian analysis (BA) was performed using MrBayes version 3.2.6 (Ronquist et al. 2012) for two million generations with the selected model. Phylogenetic trees were viewed and edited in FigTree version 1.4.2. Alignment and phylogenetic trees from all methods have been deposited in TreeBASE (S19303).
Ancestral character state reconstructions of sporangial papillation
Information on the sporangial papillation of individual species was compiled from the literature (Erwin & Ribeiro 1996, Gallegly & Hong 2008, Kroon et al. 2012, Martin et al. 2012) with emphasis given to their respective original descriptions (Table 1). Both likelihood and parsimony ancestral state reconstructions were performed on the ML tree from the phylogenetic analyses using Mesquite version 3.03 (Maddison & Maddison 2017).
RESULTS
Sequences, alignment, and phylogenetic model
PCR amplification and sequencing was successful for almost all isolates and seven genetic markers. Failure to obtain sequences only occurred occasionally for a few isolates, such as the EF1α gene of Phytophthora bilorbang (61G8), the Enl gene of P. macrochlamydospora (33E1, 31E9, and 33D5), and P. quininea (45F2), and TigA of P. megasperma (62C7) (Supplementary Table 1). These failures were set as missing data in the alignment. After trimming, each isolate was represented by an 8435-bp concatenated sequence in the alignment including gaps and missing data. This included 496 bp for 60S, 1136 bp for Btub, 965 bp for EF1α, 1169 bp for Enl, 1758 bp for HSP90, 1270 bp for 28S, and 1641 bp for TigA (TreeBASE S19303). The general time reversible nucleotide substitution model with gamma-distributed rate variation and a proportion of invariable sites (GTR+I+G) was identified by jModelTest as the most appropriate model for the phylogenetic analyses.
An expanded phylogeny including 10 clades and basal taxa
The three phylogenetic analysis methods, including ML, MP, and BA analyses (TreeBASE S19303), resulted in similar tree topologies. The topology and branch lengths of the ML inference are shown in Fig. 1. The monophyly of each of the previously recognized 10 clades was generally well supported with a few exceptions. Specifically, all clades except for clade 4 were highly supported by > 95 % bootstrap values in ML analysis and 100 % posterior probability (PP) in BA analysis (Fig. 1). Clades 1–3, 5, 7, and 10 were also highly supported by > 95 % bootstrap values in the MP analysis (Fig. 1). However, clades 6, 8, and 9, were only moderately supported with bootstrap numbers of 68, 61, and 52 in the MP analysis, respectively (Fig. 1).
Fig. 1.
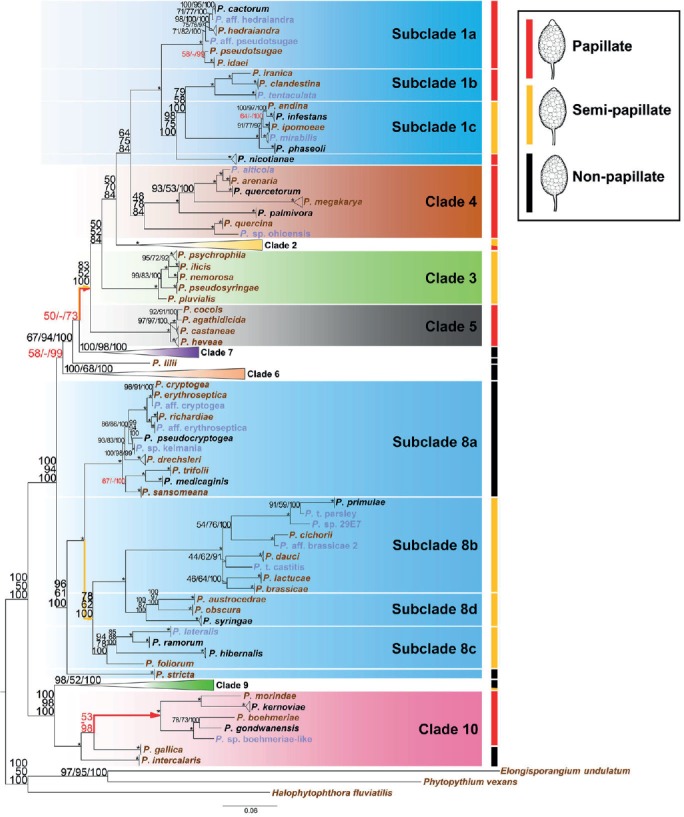
A phylogeny for the genus Phytophthora based on concatenated sequences of seven nuclear genetic markers. Topology and branch lengths of maximum likelihood analysis are shown. Bootstrap values for maximum likelihood and maximum parsimony, and Bayesian posterior probabilities (percentages) are indicated on individual nodes and separated by a forward slash. An asterisk is used in place of nodes with unambiguous (100 %) support in all three analyses. A dash is used in place of a topology from an analysis ambiguous to the other two analyses and these sets of numbers with ambiguity in one analysis are also highlighted in red. Detailed structures of clades 2, 6, 7, and 9 are shown in Fig. 2, 3, 4, 5, respectively. Species represented by ex-types and authentic isolates are written in brown and blue, respectively. Branches indicating three hypothesized evolutionary paths with all species producing papillate or semi-papillate sporangia are drawn in red or orange, respectively. Scale bar indicates number of substitutions per site.
As nearly half of all taxa included in this phylogeny were recently described, all clades in this phylogeny are expanded here to various extents compared to previously published phylogenies. The general structure of clades 1, 3, 5, 8 and 10 remained as previously assigned by Blair et al. (2008) and Martin et al. (2014) with additions of new species. For example, clade 1 was divided into three well-supported subclades and P. nicotianae was placed basal to subclades 1b and 1c (Fig. 1). Clade 8 was divided into four generally well-supported subclades, except P. stricta, which was placed basal to all clade 8 species (Fig. 1). New subclades were assigned to clade 2 (Fig. 2), clade 6 (Fig. 3), clade 7 (Fig. 4) and clade 9 (Fig. 5).
Fig. 2.
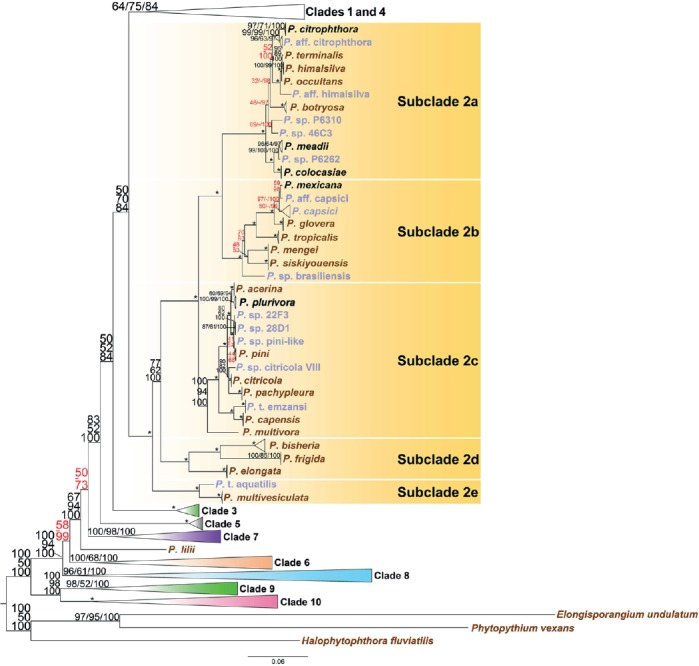
Structure of Phytophthora clade 2 in a genus-wide phylogeny for the genus Phytophthora based on concatenated sequences of seven nuclear genetic markers. Topology and branch lengths of maximum likelihood analysis are shown. Bootstrap values for maximum likelihood and maximum parsimony, and Bayesian posterior probabilities (percentages) are indicated on individual nodes and separated by a forward slash. An asterisk is used in place of nodes with unambiguous (100 %) support in all three analyses. A dash is used in place of a topology from an analysis ambiguous to the other two analyses and these sets of numbers with ambiguity in one analysis are also highlighted in red. Species represented by ex-types and authentic isolates are written in brown and blue, respectively. Scale bar indicates number of substitutions per site.
Fig. 3.
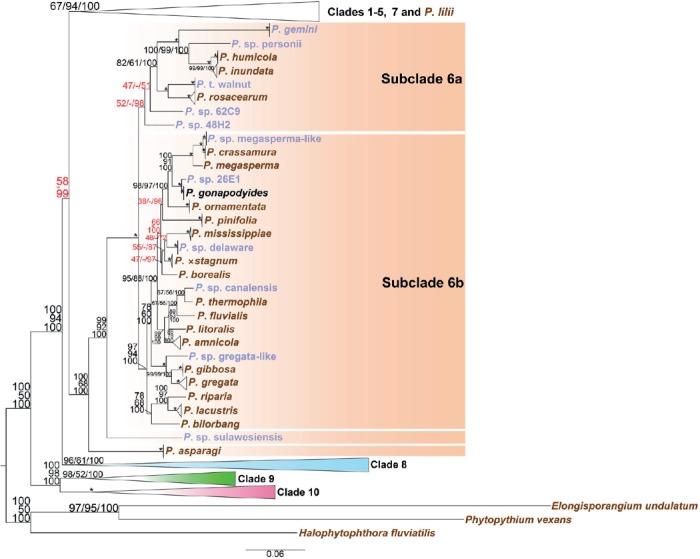
Structure of Phytophthora clade 6 in a genus-wide phylogeny for the genus Phytophthora based on concatenated sequences of seven nuclear genetic markers. Topology and branch lengths of maximum likelihood analysis are shown. Bootstrap values for maximum likelihood and maximum parsimony, and Bayesian posterior probabilities (percentages) are indicated on individual nodes and separated by a forward slash. An asterisk is used in place of nodes with unambiguous (100 %) support in all three analyses. A dash is used in place of a topology from an analysis ambiguous to the other two analyses and these sets of numbers with ambiguity in one analysis are also highlighted in red. Species represented by ex-types and authentic isolates are written in brown and blue, respectively. Scale bar indicates number of substitutions per site.
Fig. 4.
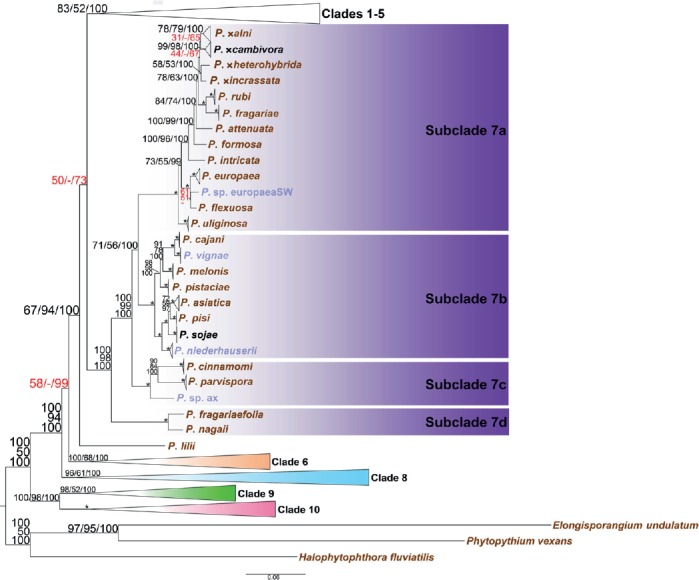
Structure of Phytophthora clade 7 in a genus-wide phylogeny for the genus Phytophthora based on concatenated sequences of seven nuclear genetic markers. Topology and branch lengths of maximum likelihood analysis are shown. Bootstrap values for maximum likelihood and maximum parsimony, and Bayesian posterior probabilities (percentages) are indicated on individual nodes and separated by a forward slash. An asterisk is used in place of nodes with unambiguous (100 %) support in all three analyses. A dash is used in place of a topology from an analysis ambiguous to the other two analyses and these sets of numbers with ambiguity in one analysis are also highlighted in red. Species represented by ex-types and authentic isolates are written in brown and blue, respectively. Scale bar indicates number of substitutions per site.
Fig. 5.
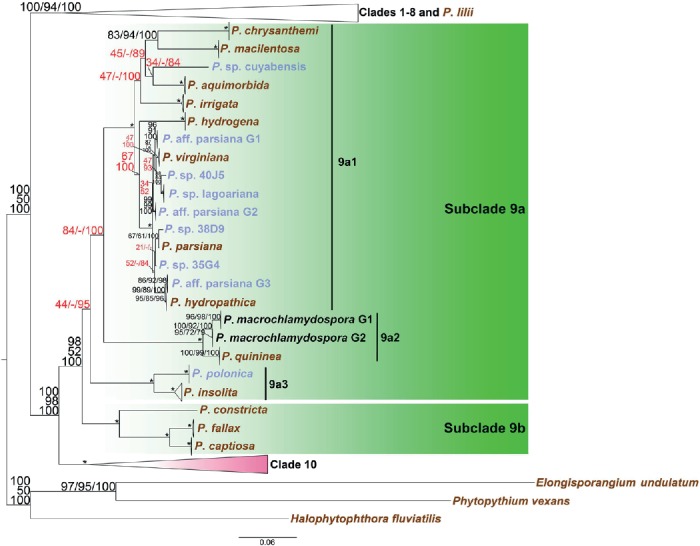
Structure of Phytophthora clade 9 in a genus-wide phylogeny for the genus Phytophthora based on concatenated sequences of seven nuclear genetic markers. Topology and branch lengths of maximum likelihood analysis are shown. Bootstrap values for maximum likelihood and maximum parsimony, and Bayesian posterior probabilities (percentages) are indicated on individual nodes and separated by a forward slash. An asterisk is used in place of nodes with unambiguous (100 %) support in all three analyses. A dash is used in place of a topology from an analysis ambiguous to the other two analyses and these sets of numbers with ambiguity in one analysis are also highlighted in red. Species represented by ex-types and authentic isolates are written in brown and blue, respectively. Scale bar indicates number of substitutions per site.
Several species were placed basal to other species in their respective clades. First, the cluster of P. quercina and P. sp. ohioensis was placed basal to other species of clade 4 in all three analyses. The bootstrap supports of the ML and MP analyses, and PP (percentage) for the separation of this cluster from that of P. alticola, P. arenaria, P. megakarya, P. palmivora, and P. quercetorum in clade 4 were only 48, 78, and 84, respectively (Fig. 1). Second, P. lilii was excluded from all known clades; it was placed basal to clades 1–5 and 7 (Fig. 1). Third, in clade 6, bootstrap support for the ML and MP analyses, and PP for all species except P. asparagi and P. sp. sulawesiensis were 100/100/100 (Fig. 3). This set of support numbers decreased to 99/92/100 when P. sp. sulawesiensis was included, and to 100/68/100 when further including P. asparagi (Fig. 3). Fourth, the support numbers for clade 8 species excluding P. stricta was 100/100/100, but 96/61/100 when P. stricta was included (Fig. 1). Fifth, all papillate species in clade 10 (Table 1) formed a well-supported main cluster, while two more recently described non-papillate species, P. gallica and P. intercalaris, were placed basal to the main cluster (Fig. 1).
New subclades in clades 2, 6, 7, and 9
(a) Clade 2
In addition to the previously recognized subclades 2a and 2b, many species, such as P. acerina, P. capensis, P. citricola, P. multivora, P. pachypleura, P. plurivora, and P. pini in the commonly referred to “Phytophthora citricola-complex” defined a new subclade 2c (Fig. 2). Furthermore, P. bisheria, P. frigida, and P. elongata formed new subclade 2d and the cluster of P. multivesiculata and P. taxon aquatilis formed new subclade 2e, with maximum support values in each case (Fig. 2).
(b) Clade 6
Subclade 6a included P. gemini, P. humicola, P. inundata, P. rosacearum, P. sp. personii, P. sp. 48H2, P. sp. 62C9 and P. taxon walnut. The cluster of P. rosacearum and P. taxon walnut could not be separated from that represented by P. gemini with only moderate support values for separation (82/61/100) (Fig. 3). Isolates 62C9 and 48H2, belonging to two new species, had ambiguous placements within subclade 6a among the three analyses (Fig. 3). With approximately 20 species newly included in the present phylogeny, the previously recognized “P. megasperma-P. gonapodyides complex” (Brasier et al. 2003a), subclade II of clade 6 (Jung et al. 2011), or subclade 6b (Kroon et al. 2012) expanded and its separation from subclade 6a was well-supported by 100/100/100 values (Fig. 3). Within subclade 6b, separation of the cluster of P. bilorbang, P. lacustris, and P. riparia from the other subclade 6b species was highly supported by 97/94/100 (Fig. 3), indicating that these three species may define a new subclade, although this is not done in this study. Phytophthora sp. sulawesiensis was placed basal to other clade 6 species except for P. asparagi, while P. asparagi was basal to all other species in clade 6 (Fig. 3). Phytophthora asparagi was previously assigned as subclade 6c (Kroon et al. 2012) and subclade III of clade 6 (Jung et al. 2011); considering that the support value of MP analysis was only moderate (68 %) when this single taxon was included (Fig. 3), this previous assignation as a subclade was not adopted here. In addition, in order to be consistent with subclade names in other clades, subclades 6a and 6b were used here instead of subclades I and II by Jung et al. (2011).
(c) Clade 7
Four subclades were distinguished in clade 7. Separation of the previously assigned subclades 7a and 7b was only moderately supported by values 71/56/100 (Fig. 4). The general structure of subclade 7a remained the same even with the addition of seven new taxa. Six of these new species, including P. attenuata, P. flexuosa, P. formosa, P. intricata, P. ×heterohybrida, and P. ×incrassata were recently recovered from forest soils and streamwater in Taiwan (Jung et al. 2017). On the other hand, P. cinnamomi and P. parvispora were separated from subclade 7b. They, along with a provisional species, P. sp. ax from Virginia, USA (Table 1), formed a distinct new subclade 7c (Fig. 4). The new subclade 7d, including two recently described species from Japan (Rahman et al. 2014b), P. fragariaefolia and P. nagaii, was placed basal to other subclades in clade 7 (Fig. 4).
(d) Clade 9
The split of clade 9 into two subclades 9a and 9b was highly supported in ML (98 %) and BA (100 %) analyses and moderately supported in the MP (52 %) analysis (Fig. 5). However, monophyly was highly supported for subclade 9b (100/100/100) but not for subclade 9a (44/-/95) (Fig. 5). Within subclade 9a, three monophyletic clusters were formed: 9a1, 9a2, and 9a3. However, support for the separation of these three clusters was moderate or ambiguous. In particular, the MP results did not produce any consistent separation of the three clusters (Fig. 5). Cluster 9a1 included many recently described high-temperature tolerant species, such as P. aquimorbida, P. chrysanthemi, P. hydropathica, P. macilentosa, P. parsiana, and P. virginiana). The cluster of P. macrochlamydospora (two lineages with two isolates in each lineage, Table 1) and P. quininea constituted 9a2 (Fig. 5). The cluster of two other high-temperature tolerant species P. insolita and P. polonica constituted 9a3 (Fig. 5). The well-supported cluster of P. captiosa, P. constricta, and P. fallax was assigned as subclade 9b (Fig. 5).
Evolutionary history of sporangial papillation inferred from ancestral character state reconstructions
Sporangial papillation of individual species is indicated in Table 1 and Fig. 6. Due to the size of the cladograms, clusters including species with the same sporangial papillation within each (sub)clade were compressed in Mesquite. Both likelihood and parsimony methods suggested that non-papillate is the progenitor state of Phytophthora species, and that semi-papillate and papillate types were derived from the non-papillate. The analyses indicated three major clusters of semi-papillate and (or) papillate species diverged from the non-papillate ancestors. First, species in clades 1 to 5 (semi-papillate or papillate) diverged from non-papillate species in clade 7 and P. lilii (Fig. 6). Second, species in subclades 8b to 8d (semi-papillate) diverged from non-papillate subclade 8a species (Fig. 6). Third, papillate clade 10 species including P. boehmeriae, P. gondwanensis, P. kernoviae, and P. morindae diverged from the non-papillate P. gallica and P. intercalaris (Fig. 6). Several species such as P. macrochlamydospora, P. mississippiae, P. gibbosa, and P. constricta also evolved to produce partially semi-papillate sporangia (Fig. 6).
Fig. 6.
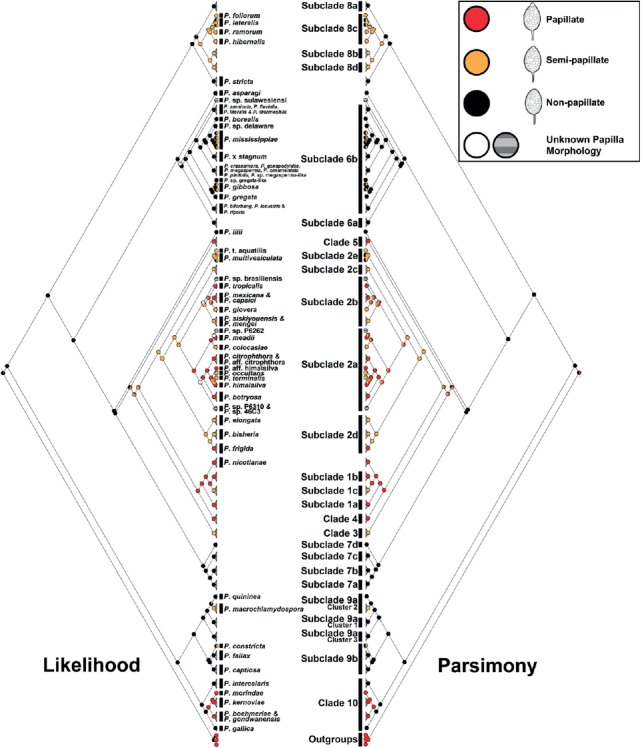
Ancestral state reconstructions of sporangial papillation for the genus Phytophthora based on likelihood (left cladogram) and parsimony (right cladogram). Trace character history analyses were performed on the maximum likelihood phylogeny in Mesquite. Clusters including species of uniform sporangial papillation within individual (sub)clades were compressed in Mesquite.
DISCUSSION
Here we presented an expanded phylogeny for the genus Phytophthora, encompassing 142 formally named and 43 provisionally recognized species (Table 2). In addition to this comprehensive coverage, this expanded phylogeny features over 1500 signature sequences generated from 278 ex-type and authentic isolates of 162 Phytophthora taxa (Supplementary Table 1). Furthermore, this study provided new insights into the evolutionary history of sporangial papillation in Phytophthora.
The expanded phylogeny provides a sound taxonomic framework for this agriculturally and ecologically important genus. One hundred and fourteen ex-types were included, representing 80 % of the 142 formally named species in this phylogeny. The majority of the 29 species not represented by ex-types, such as P. gonapodyides, P. infestans, P. meadii, P. mexicana, and P. nicotianae, were described long ago without designation of an ex-type culture. Likewise, almost all the 43 provisional species in this phylogeny were represented by authentic isolates from the originators of the respective species (Table 1 and Supplementary Table 1). This new framework will facilitate identification of new taxa in the future. As the genus continues to rapidly expand, some recently described species were not included in this study: P. mekongensis in subclade 2a (Puglisi et al. 2017), P. amaranthi in subclade 2b (Ann et al. 2016), P. boodjera in clade 4 (Simamora et al. 2015), P. chlamydospora in subclade 6b (Hansen et al. 2015), P. uniformis (basionym: P. alni subsp. uniformis) and P. ×multiformis (basionym: P. alni subsp. multiformis) in subclade 7a (Brasier et al. 2004, Husson et al. 2015), P. pseudolactucae in subclade 8b (Rahman et al. 2015), and P. prodigiosa (Puglisi et al. 2017) and P. pseudopolonica (Li et al. 2017) in subclade 9a. Likewise, some informally designated species also were not included: such as P. taxon humicola-like, P. taxon kwongan, and P. taxon rosacearum-like in subclade 6a (Jung et al. 2011). These and other emerging species are yet to be incorporated in the overall phylogeny of the genus.
The generation of over 1500 signature sequences from ex-types and authentic isolates in this study will aid researchers and first responders in correctly identifying Phytophthora cultures to the species level. DNA sequencing of selected genetic markers has become common practice in the identification of Phytophthora cultures (Kang et al. 2010). However, it is recognized that the accuracy of culture identity determined by this approach depends on the quality of the reference sequences used – and currently many sequence deposits are erroneously identified in public repositories, including GenBank (Kang et al. 2010). These errors originated in sequence deposits of cultures that were identified by morphological characters alone, and compounded by those identified through sequence matches to erroneous reference sequences or by single DNA markers (Kang et al. 2010). In this study, 29 isolates were found associated with an erroneous or modified identity (Supplementary Table 2). For instance, isolate 29B3 in clade 1 was identified as P. pseudotsugae and used as a key isolate for this species by Gallegly & Hong (2008). However, its sequences were distinct from those of the P. pseudotsugae ex-type (ATCC 52938). In the phylogenetic tree, it was basal to the cluster of P. cactorum and P. hedraiandra, thus its species identity was changed to P. aff. pseudotsugae (Fig. 1). In clade 2, isolate 26H4 was identified as P. citrophthora (Gallegly & Hong 2008) but sequences and phylogeny showed that it was close to but distinct from P. citrophthora isolates 03E5 and 26H3. It formed a cluster with isolate IMI 342898 (P10341), which was coded as P. sp. aff. colocasiae-1 by Martin et al. (2014). The identity of both isolates was then changed to P. aff. citrophthora (Fig. 2). Similarly, in clade 8, isolate 22G2 had been identified as P. cryptogea, although it was distinct from the P. cryptogea ex-type 61H9 (CBS 113.19). In the phylogenetic tree, it was basal to the cluster of P. cryptogea and P. erythroseptica, and the species identity was consequently changed to P. aff. cryptogea (Fig. 1). Changes in the identifications of these isolates, including the new and original names used, are indicated in Supplementary Table 2. The changes in the naming of these isolates highlights the importance of using signature sequences from ex-type or authentic isolates as references in future culture identification. In order to facilitate this practice, the signature sequences generated from ex-types or authentic isolates in the present study are marked as ‘(ex-type)’ or ‘(authentic)’, respectively, under the ‘isolate’ section in the ‘feature’ table of GenBank deposits. The research, diagnostic and regulatory communities are encouraged to use these sequences as references in future culture identification.
This study provided new insights into the evolutionary history of sporangial morphology in the genus Phytophthora, a subject that has fascinated generations of mycologists and plant pathologists. There have been three major hypotheses regarding the development of papillation, as illustrated in Fig. 7a, b, and c, respectively. First, papillate species were considered as descendants of Pythium-like, non-papillate ancestors and semi-papillation has been considered as intermediate between non-papillation and papillation (Blackwell 1949, Cooke et al. 2000, Erwin & Ribeiro 1996). Second, some semi-papillate species, exemplified by P. primulae in the group III of Waterhouse (1963) are primitive; they were suggested to have evolved to papillate and non-papillate species through two distinct evolutionary lines (Brasier 1983). Third, semi-papillate sporangia are morphological variants of papillate and non-papillate types (Cooke et al. 2000). Here we suggest that the non-papillate type is ancestral, and that non-papillate species could have evolved directly into either semi-papillate or papillate species (Fig. 7d). The evolution to semi-papillate species is exemplified by those in subclades 8b–d (Fig. 1), while evolution to papillate species is illustrated by P. boehmeriae and other papillate species in clade 10 (Fig. 1).The relationship between semi-papillate and papillate species appears to be more complicated (Fig. 7d). We also hypothesize that some semi-papillate species, such as those in subclade 1c, may have diverged from papillate ancestors, while some papillate species such as P. frigida may have evolved from semi-papillate ancestors of subclade 2d (Fig. 6).
Fig. 7.
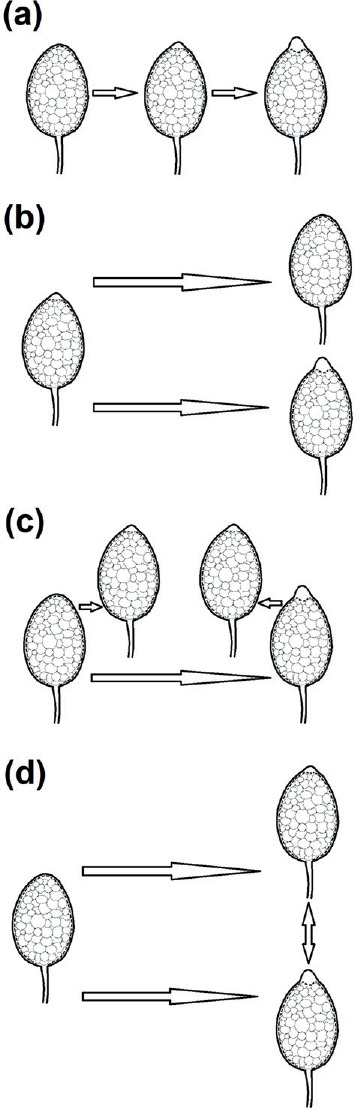
Illustration of hypotheses on evolution of Phytophthora and associated changes in sporangial papillation: (a) species producing papillate sporangia evolved from non-papillate ancestors. Semi-papillation is considered as intermediate between non-papillation and papillation (Blackwell 1949, Cooke et al. 2000, Erwin & Ribeiro 1996); (b) some semi-papillate species, exemplified by P. primulae in the group III of Waterhouse (1963), are primitive and evolved to be non-papillate and papillate through two evolutionary paths, by Brasier (1983); (c) papillate species evolved from non-papillate ancestors. Semi-papillate species have been considered as morphological variants of papillate or non-papillate species, by Cooke et al. (2000); (d) a new hypothesis developed in this study that non-papillate ancestors evolved directly to either papillate or semi-papillate species. Some semi-papillate species further evolved to be papillate, or vice versa.
These new hypotheses are supported by the results from phylogeny and ancestral state reconstructions that suggest three major evolutionary paths in sporangial papillation of Phytophthora species (Fig. 1). First, the ancestor of modern species in clades 1–5 evolved to be papillate or semi-papillate (Figs 1, 6) while diverging from the common non-papillate ancestor of clade 7 species (Figs 1, 6). Second, the common ancestor of species in subclades 8b–d diverged from that of subclade 8a species while acquiring semi-papillation (Figs 1, 6). Third, the common ancestor of five clade 10 species in the main cluster including P. boehmeriae, P. gondwanensis, P. kernoviae, P. morindae, and P. sp. boehmeriae-like, acquired papillate sporangia while diverging from two non-papillate clade 10 species, P. gallica and P. intercalaris (Figs 1, 6). Besides these three major groups of papillate or semi-papillate species, a few species may have evolved to acquire semi-papillation independently, such as P. macrochlamydospora in clade 9 (Fig. 6). This evolutionary process may be underway for some other species including P. constricta, P. gibbosa, and P. mississippiae, which all produce both semi-papillate and non-papillate sporangia (Fig. 6). Furthermore, evolutionary reversion to partial production of non-papillate sporangia may have occurred in P. multivesiculata and P. lateralis in two semi-papillate subclades 2e and 8c, respectively (Fig. 6). However, that conclusion is uncertain due to limited and ambiguous data from species in these two subclades. Specifically, P. lateralis was ambiguously reported as non-papillate (Erwin & Ribeiro 1996, Gallegly & Hong 2008, Martin et al. 2012, Tucker & Milbrath 1942) or non- to semi-papillate (Kroon et al. 2012) in different studies. In subclade 2e, the only sister taxon of P. multivesiculata, P. taxon aquatilis, was provisionally described as semi-papillate, but only based on a single isolate (Hong et al. 2012). Evolutionary reversion in the sporangial papillation of these two species requires validation in the future. Also, more studies are warranted to analyze additional characters based on phylogenies with better clade-to-clade resolutions and provide a more comprehensive picture on the evolutionary history of Phytophthora species.
That a number of species were placed basal to other species in their respective clades in this expanded phylogeny presents a significant challenge to the monophyly of their respective clades and the current 10-clade system. First, P. stricta was initially placed close to other species in subclade 8a based on sequences of the cytochrome c oxidase 1 (cox1) gene, but was not grouped in any ITS clade (Yang et al. 2014a). This species was grouped in clade 8 in our expanded phylogeny by ML and BA analyses (Fig. 1); the monophyly of this clade was only moderately supported (61 %) in the MP analysis (Fig. 1). Second, the monophyly of clade 6 including P. asparagi was only moderately supported (68 %) in the MP analysis (Fig. 3). Third, although the inclusion of P. intercalaris in clade 10 was supported with maximum values, the exact positions of this species and P. gallica were still unresolved since the next node was only moderately supported (53 %) in the ML analysis and ambiguous in the MP analysis (Fig. 1). Fourth, similar to the finding of Blair et al. (2008), support for the monophyly of clade 4 including P. quercina and P. sp. ohioensis was only moderate (48/78/84). Also, similar ambiguity in the placement of the ‘P. quercina – P. sp. ohioensis’ cluster was observed among different phylogenetic approaches, and using different datasets including nuclear, mitochondrial, and combined nuclear and mitochondrial sequences (Martin et al. 2014). Fifth, this phylogeny confirmed the finding by Rahman et al. (2015) that P. lilii was not grouped in any clade of the current 10-clade system (Fig. 1). This species was not assigned as a distinct clade in our study, due to the relatively low clade-to-clade resolutions (Fig. 1). Further analyses are warranted to determine whether this unique species should be assigned as a new clade.
Although many branches in the expanded phylogeny have consistent maximum support in all three methods, some have only moderate to low or inconsistent support. These results highlight the challenges of correctly inferring the evolutionary separation of many closely related Phytophthora species, even when concatenated sequences from seven phylogenetic markers were used. It can be expected that as the cost of gene sequencing drops further, it will become possible to increase phylogenetic resolution among Phytophthora species by using concatenations of much larger numbers of genes. For example, Ye et al. (2016) used 293 concatenated housekeeping proteins to infer a robust phylogeny of seven fully sequenced Phytophthora species and confirmed that downy mildews (represented by three genome sequences) are nested within the genus Phytophthora, close to Phytophthora clade 4 (Ye et al. 2016). However, even with full genome sequences, ambiguity may not be completely resolved in cases where speciation has involved large populations of sexually reproducing individuals, for example, as a result of geographic separation. In these cases, there may be many sequence polymorphisms shared among separated species and these may confound the inference of a reliable phylogeny. Resolution of this level of ambiguity may require sequencing the whole genome of many isolates from the species of interest as well as using improved phylogenetic and coalescent methods.
With the number of described Phytophthora species increasing, recent studies have raised an important concern in the accurate detection of species boundaries using phylogenetic data (Jung & Burgess 2009, Pánek et al. 2016, Safaiefarahani et al. 2015). One example is the status of P. hedraiandra as a distinct species in subclade 1a (Pánek et al. 2016). As evidenced by the amplified fragment length polymorphism (AFLP) and phylogenetic analysis based on sequences of ITS, phenolic acid decarboxylase, and cox1 genes, a recent study concluded that P. hedraiandra was just one lineage of P. cactorum, while morphological data provided only limited information to delimitate these two species (Pánek et al. 2016). Also, phylogenetic analyses in this study indicated that P. cactorum and P. hedraiandra cluster with strong support (98/100/100), and P. aff. hedraiandra isolate 33F4 (previously identified as P. hedraiandra Supplementary Table 2), was clustered with P. cactorum (Fig. 1). Phylogenies based on nuclear sequences prior to this study also supported P. hedraiandra as closely related to P. cactorum (Blair et al. 2008, Martin et al. 2014). However, in the phylogenies based on concatenated sequences of four mitochondrial loci, and combined seven nuclear and four mitochondrial loci, P. hedraiandra was basal to the cluster of P. cactorum and P. pseudotsugae, and clustered with P. idaei, respectively (Martin et al. 2014). Phytophthora cactorum and P. hedraiandra also have very distinctive single-strand-conformation polymorphism patterns (Gallegly & Hong 2008). Apparently, more investigations are warranted to resolve the P. cactorum complex. Likewise, indistinct boundaries are present among species in other subclades, such as the ‘P. citricola complex’ or subclade 2c (Brazee et al. 2017, Jung & Burgess 2009), the ‘P. cryptogea complex’ in subclade 8a (Safaiefarahani et al. 2015, 2016) and cluster 9a1 in subclade 9a including P. hydropathica (Hong et al. 2010), P. parsiana (Mostowfizadeh-Ghalamfarsa et al. 2008), P. virginiana (Yang & Hong 2013) and other provisionally designated species. Accurately delimiting these closely related species within the genus remains an important task.
This expanded phylogeny has highlighted the importance and difficulty of accurately interpreting the position of hybrid Phytophthora species. As exemplified by P. ×alni (Brasier et al. 2004, Husson et al. 2015), many hybrid species have been identified among emerging plant pathogens (Jung et al. 2017, Man in’t Veld et al. 2012, Nirenberg et al. 2009). Due to the presence of multiple alleles originated from parent species in their nuclear genes, phylogenetic analysis of these hybrids based on nuclear sequences alone may not produce a robust placement. As illustrated in this phylogeny, the placement of hybrid species may be ambiguous. Specifically, in subclade 6b, support values for the placement of P. ×stagnum and its closely related species, P. mississippiae, P. borealis, and P. sp. delaware were moderate in the ML and BA analyses and ambiguous in the MP analysis (Fig. 3). Similarly, in subclade 7a, the placement of P. ×alni, P. ×cambivora, P. ×heterohybrida, and P. ×incrassata’ cluster was not well resolved due to ambiguous placement in the MP analysis and moderate support values in the other two analyses (Fig. 4). Adding mitochondrial sequences into the phylogenetic analyses may be a solution to this problem. However, due to the uniparental inheritance of mitochondria, the hybrids and their maternal parents are inseparable by mitochondrial sequences and their placements could conflict with nuclear analyses (Martin et al. 2014).
Acknowledgments
This research was supported in part by grants from the USDA-NIFA-Specialty Crop Research Initiative (Agreement no. 2010-51181-21140). We would like to thank all authorities and species originators who provided Phytophthora isolates to our study, including Yilmaz Balci, Zia Banihashemi, Lien Bertier, Karien Bezuidenhout, Clive Brasier, Treena Burgess, Mike Coffey, Mannon Gallegly, Beatrice Ginetti, Niklaus Grünwald, Everett Hansen, Beatrice Henricot, Fredrik Heyman, Hon Ho, Maria Holeva, Steven Jeffers, Thomas Jung, Koji Kageyama, Willem Man in 't Veld, Jan Nechwatal, Bruno Scanu, Andrea Vannini, Anna Maria Vettraino, and Irene Vloutoglou. Names of many other contributors are listed in Supplementary Table 1.
Supplementary Material
REFERENCES
- Abad ZG, Abad JA, Cacciola SO, Pane A, Faedda R, et al. (2014) Phytophthora niederhauserii sp. nov., a polyphagous species associated with ornamentals, fruit trees and native plants in 13 countries. Mycologia 106: 431–437. [DOI] [PubMed] [Google Scholar]
- Abad ZG, Abad JA, Coffey MD, Oudemans PV, Man WA, et al. (2008) Phytophthora bisheria sp. nov., a new species identified in isolates from the Rosaceous raspberry, rose and strawberry in three continents. Mycologia 100: 99–110. [DOI] [PubMed] [Google Scholar]
- Abad ZG, Ivors KL, Gallup CA, Abad JA, Shew HD. (2011) Morphological and molecular characterization of Phytophthora glovera sp. nov. from tobacco in Brazil. Mycologia 103: 341–350. [DOI] [PubMed] [Google Scholar]
- Aghighi S, Hardy GESJ, Scott JK, Burgess TI. (2012) Phytophthora bilorbang sp. nov., a new species associated with the decline of Rubus anglocandicans (European blackberry) in Western Australia. European Journal of Plant Pathology 133: 841–855. [Google Scholar]
- Amin KS, Baldev B, Williams FJ. (1978) Phytophthora cajani, a new species causing stem blight on Cajanus cajan. Mycologia 70: 171–176. [Google Scholar]
- Ann PJ, Huang JH, Tsai JN, Ko WH. (2016) Morphological, molecular and pathological characterization of Phytophthora amaranthi sp. nov. from Amaranth in Taiwan. Journal of Phytopathology 164: 94–101. [Google Scholar]
- Ann PJ, Ko WH. (1980) Phytophthora insolita, a new species from Taiwan. Mycologia 72: 1180–1185. [Google Scholar]
- Aragaki M, Uchida JY. (2001) Morphological distinctions between Phytophthora capsici and P. tropicalis sp. nov. Mycologia 93: 137–145. [Google Scholar]
- Balci Y, Balci S, Blair JE, Park SY, Kang S, et al. (2008) Phytophthora quercetorum sp. nov., a novel species isolated from eastern and north-central USA oak forest soils. Mycological Research 112: 906–916. [DOI] [PubMed] [Google Scholar]
- Belbahri L, Moralejo E, Calmin G, Oszako T, Garcia JA, et al. (2006) Phytophthora polonica, a new species isolated from declining Alnus glutinosa stands in Poland. FEMS Microbiology Letters 261: 165–174. [DOI] [PubMed] [Google Scholar]
- Bertier L, Brouwer H, De Cock A, Cooke DEL, Olsson CHB, et al. (2013) The expansion of Phytophthora clade 8b: three new species associated with winter grown vegetable crops. Persoonia 31: 63–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezuidenhout CM, Denman S, Kirk SA, Botha WJ, Mostert L, et al. (2010) Phytophthora taxa associated with cultivated Agathosma, with emphasis on the P. citricola complex and P. capensis sp. nov. Persoonia 25: 32–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell E. (1949) Terminology in Phytophthora. Mycological Papers 30: 1–24. [Google Scholar]
- Blair JE, Coffey MD, Park S-Y, Geiser DM, Kang S. (2008) A multi-locus phylogeny for Phytophthora utilizing markers derived from complete genome sequences. Fungal Genetics and Biology 45: 266–277. [DOI] [PubMed] [Google Scholar]
- Brasier CM. (1983) Problems and prospects in Phytophthora research. In: Phytophthora: its Biology, taxonomy, ecology, and pathology (Erwin DC, Bartnicki-Garcia S, Tsao PH, eds): 351–364. St Paul, MN: American Phytopathological Society Press. [Google Scholar]
- Brasier CM, Beales PA, Kirk SA, Denman S, Rose J. (2005) Phytophthora kernoviae sp. nov., an invasive pathogen causing bleeding stem lesions on forest trees and foliar necrosis of ornamentals in the UK. Mycological Research 109: 853–859. [DOI] [PubMed] [Google Scholar]
- Brasier CM, Cooke DEL, Duncan JM, Hansen EM. (2003a) Multiple new phenotypic taxa from trees and riparian ecosystems in Phytophthora gonapodyides–P. megasperma ITS Clade 6, which tend to be high-temperature tolerant and either inbreeding or sterile. Mycological Research 107: 277–290. [DOI] [PubMed] [Google Scholar]
- Brasier CM, Griffin MJ. (1979) Taxonomy of ‘Phytophthora palmivora’ on cocoa. Transactions of the British Mycological Society 72: 111–143. [Google Scholar]
- Brasier CM, Kirk SA, Delcan J, Cooke DEL, Jung T, et al. (2004) Phytophthora alni sp. nov. and its variants: designation of emerging heteroploid hybrid pathogens spreading on Alnus trees. Mycological Research 108: 1172–1184. [DOI] [PubMed] [Google Scholar]
- Brasier CM, Sanchez-Hernandez E, Kirk SA. (2003b) Phytophthora inundata sp. nov., a part heterothallic pathogen of trees and shrubs in wet or flooded soils. Mycological Research 107: 477–484. [DOI] [PubMed] [Google Scholar]
- Brazee NJ, Yang X, Hong C. (2017) Phytophthora caryae sp. nov., a new species recovered from streams and rivers in the eastern United States. Plant Pathology 66: 805–817. [Google Scholar]
- Breda De Haan JV. (1896) De bibitziekte in de Deli-tabak veroorzaakt door Phytophthora nicotianae. Mededeelingen uit ‘s Lands Plantentuin Batavia 15: 1–107. [Google Scholar]
- Buddenhagen IW, Young RA. (1957) A leaf and twig disease of English holly caused by Phytophthora ilicis n. sp. Phytopathology 47: 95–101. [Google Scholar]
- Buisman CJ. (1927) Root rots caused by Phycomycetes. Mededelingen Phytopathologisch Laboratorium “Willie Commelin Scholten” 11:1–51. [Google Scholar]
- Butler EJ. (1910) The bud-rot of palms in India. Memoirs of the Department of Agriculture in India, Botanical Series 3: 221–280. [Google Scholar]
- Carne WM. (1925) A brown rot of citrus in Australia (Phytophthora hibernalis n. sp). Journal of the Royal Society of Western Australia 12: 13–41. [Google Scholar]
- Chee KH. (1969) Variability of Phytophthora species from Hevea brasiliensis. Transactions of the British Mycological Society 52: 425–436. [Google Scholar]
- Cooke DEL, Drenth A, Duncan JM, Wagels G, Brasier CM. (2000) A molecular phylogeny of Phytophthora and related oomycetes. Fungal Genetics and Biology 30: 17–32. [DOI] [PubMed] [Google Scholar]
- Crandall BS. (1947) A new Phytophthora causing root and collar rot of Cinchona in Peru. Mycologia 39: 218–223. [PubMed] [Google Scholar]
- Crous PW, Groenewald JZ, Shivas RG, Edwards J, Seifert KA, et al. (2011) Fungal Planet description sheets: 69–91. Persoonia 26: 108–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Summerell BA, Shivas RG, Burgess TI, Decock CA, et al. (2012) Fungal Planet description sheets: 107–127. Persoonia 28: 138–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Le Roux JJ, Richardson DM, Strasberg D, et al. (2015) Fungal Planet description sheets: 371–399. Persoonia 35: 264–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bary A. (1876) Researches into the nature of the potato-fungus, Phytophthora infestans. Journal of the Royal Agricultural Society of England 12: 239–269. [Google Scholar]
- De Cock AW, Lévesque CA. (2004) New species of Pythium and Phytophthora. Studies in Mycology 50: 481–487. [Google Scholar]
- De Cock AWAM, Lodhi AM, Rintoul TL, Bala K, Robideau GP, et al. (2015) Phytopythium: molecular phylogeny and systematics. Persoonia 34: 25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick MA, Dobbie K, Cooke DEL, Brasier CM. (2006) Phytophthora captiosa sp. nov. and P. fallax sp. nov. causing crown dieback of Eucalyptus in New Zealand. Mycological Research 110: 393–404. [DOI] [PubMed] [Google Scholar]
- Donahoo R, Blomquist CL, Thomas SL, Moulton JK, Cooke DEL, et al. (2006) Phytophthora foliorum sp. nov., a new species causing leaf blight of azalea. Mycological Research 110: 1309–1322. [DOI] [PubMed] [Google Scholar]
- Drechsler C. (1931) A crown-rot of hollyhocks caused by Phytophthora megasperma n. sp. Journal of the Washington Academy of Sciences 21: 513–526. [Google Scholar]
- Duran A, Gryzenhout M, Slippers B, Ahumada R, Rotella A, et al. (2008) Phytophthora pinifolia sp. nov. associated with a serious needle disease of Pinus radiata in Chile. Plant Pathology 57: 715–727. [Google Scholar]
- Ersek T, Ribeiro OK. (2010) Mini review article: an annotated list of new Phytophthora species described post 1996. Acta Phytopathologica et Entomologica Hungarica 45: 251–266. [Google Scholar]
- Ershad D. (1971) Beitrag zur Kenntnis der Phytophthora-arten in Iran und Ihrer Phytopathologischen Bedeutung. Berlin-Dahlem: Mitteilungen aus der Biologischen Bundesanstalt fur Land- and Forstwirtschaft. [Google Scholar]
- Erwin DC, Ribeiro OK. (1996) Phytophthora Diseases Worldwide. St Paul, MN: American Phytopathological Society Press. [Google Scholar]
- Flier WG, Grünwald NJ, Kroon L, Van Den Bosch TBM, Garay-Serrano E, et al. (2002) Phytophthora ipomoeae sp. nov., a new homothallic species causing leaf blight on Ipomoea longipedunculata in the Toluca Valley of central Mexico. Mycological Research 106: 848–856. [Google Scholar]
- Galindo-A J, Hohl HR. (1985) Phytophthora mirabilis, a new species of Phytophthora. Sydowia 38: 87–96. [Google Scholar]
- Gallegly ME, Hong C. (2008) Phytophthora: identifying species by morphology and DNA fingerprints. St Paul, MN: American Phytopathological Society Press. [Google Scholar]
- Ginetti B, Moricca S, Squires JN, Cooke DEL, Ragazzi A, et al. (2014) Phytophthora acerina sp. nov., a new species causing bleeding cankers and dieback of Acer pseudoplatanus trees in planted forests in northern Italy. Plant Pathology 63: 858–876. [Google Scholar]
- Goheen EM, Hansen EM, Kanaskie A, Mcwilliams MG, Osterbauer N, et al. (2002) Sudden oak death caused by Phytophthora ramorum in Oregon. Plant Disease 86: 441. [DOI] [PubMed] [Google Scholar]
- Greslebin AG, Hansen EM, Sutton W. (2007) Phytophthora austyocedrae sp. nov., a new species associated with Austyocedrus chilensis mortality in Patagonia (Argentina). Mycological Research 111: 308–316. [DOI] [PubMed] [Google Scholar]
- Grünwald NJ, Werres S, Goss EM, Taylor CR, Fieland VJ. (2012) Phytophthora obscura sp. nov., a new species of the novel Phytophthora subclade 8d. Plant Pathology 61: 610–622. [Google Scholar]
- Haas BJ, Kamoun S, Zody MC, Jiang RHY, Handsaker RE, et al. (2009) Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461: 393–398. [DOI] [PubMed] [Google Scholar]
- Hall TA. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- Hamm P, Hansen EM. (1983) Phytophthora pseudotsugae, a new species causing root rot of Douglas-fir. Canadian Journal of Botany 61: 2626–2631. [Google Scholar]
- Hansen EM, Maxwell DP. (1991) Species of the Phytophthora megasperma complex. Mycologia 83: 376–381. [DOI] [PubMed] [Google Scholar]
- Hansen EM, Reeser P, Sutton W, Brasier CM. (2015) Redesignation of Phytophthora taxon Pgchlamydo as Phytophthora chlamydospora sp. nov. North American Fungi 10: 1–14. [Google Scholar]
- Hansen EM, Reeser PW, Davidson JM, Garbelotto M, Ivors K, et al. (2003) Phytophthora nemorosa, a new species causing cankers and leaf blight of forest trees in California and Oregon, U.S.A. Mycotaxon 88: 129–138. [Google Scholar]
- Hansen EM, Reeser PW, Sutton W. (2012) Phytophthora borealis and Phytophthora riparia, new species in Phytophthora ITS Clade 6. Mycologia 104: 1133–1142. [DOI] [PubMed] [Google Scholar]
- Hansen EM, Wilcox WF, Reeser PW, Sutton W. (2009) Phytophthora rosacearum and P. sansomeana, new species segregated from the Phytophthora megasperma “complex”. Mycologia 101: 129–135. [DOI] [PubMed] [Google Scholar]
- Henricot B, Perez Sierra A, Jung T. (2014) Phytophthora pachypleura sp. nov., a new species causing root rot of Aucuba japonica and other ornamentals in the United Kingdom. Plant Pathology 63: 1095–1109. [Google Scholar]
- Heyman F, Blair JE, Persson L, Wikstrom M. (2013) Root rot of pea and faba bean in southern Sweden caused by Phytophthora pisi sp. nov. Plant Disease 97: 461–471. [DOI] [PubMed] [Google Scholar]
- Hickman CJ. (1940) The red core root disease of the strawberry caused by Phytophthora fragariae n. sp. Journal of Pomology and Horticultural Society 18: 89–118. [Google Scholar]
- Hong C, Gallegly ME, Richardson PA, Kong P. (2011) Phytophthora pini Leonian resurrected to distinct species status. Mycologia 103: 351–360. [DOI] [PubMed] [Google Scholar]
- Hong CX, Gallegly ME, Browne GT, Bhat RG, Richardson PA, et al. (2009) The avocado subgroup of Phytophthora citricola constitutes a distinct species, Phytophthora mengei sp. nov. Mycologia 101: 833–840. [DOI] [PubMed] [Google Scholar]
- Hong CX, Gallegly ME, Richardson PA, Kong P, Moorman GW. (2008) Phytophthora irrigata, a new species isolated from irrigation reservoirs and rivers in Eastern United States of America. FEMS Microbiology Letters 285: 203–211. [DOI] [PubMed] [Google Scholar]
- Hong CX, Gallegly ME, Richardson PA, Kong P, Moorman GW, et al. (2010) Phytophthora hydropathica, a new pathogen identified from irrigation water, Rhododendron catawbiense and Kalmia latifolia. Plant Pathology 59: 913–921. [Google Scholar]
- Hong CX, Richardson PA, Hao W, Ghimire SR, Kong P, et al. (2012) Phytophthora aquimorbida sp. nov. and Phytophthora taxon ‘aquatilis’ recovered from irrigation reservoirs and a stream in Virginia, USA. Mycologia 104: 1097–1108. [DOI] [PubMed] [Google Scholar]
- Hotson JW, Hartge L. (1923) A disease of tomato caused by Phytophthora mexicana sp. nov. Phytopathology 13: 520–531. [Google Scholar]
- Husson C, Aguayo J, Revellin C, Frey P, Ioos R, et al. (2015) Evidence for homoploid speciation in Phytophthora alni supports taxonomic reclassification in this species complex. Fungal Genetics and Biology 77: 12–21. [DOI] [PubMed] [Google Scholar]
- Ilieva E, Veld WaM, Veenbaas-Rijks W, Pieters R. (1998) Phytophthora multivesiculata, a new species causing rot in Cymbidium. European Journal of Plant Pathology 104: 677–684. [Google Scholar]
- Irwin JaG. (1991) Phytophthora macrochlamydospora, a new species from Australia. Mycologia 83: 517–519. [Google Scholar]
- Jung T, Burgess TI. (2009) Re-evaluation of Phytophthora citricola isolates from multiple woody hosts in Europe and North America reveals a new species, Phytophthora plurivora sp. nov. Persoonia 22: 95–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung T, Cooke DEL, Blaschke H, Duncan JM, Oßwald W. (1999) Phytophthora quercina sp. nov., causing root rot of European oaks. Mycological Research 103: 785–798. [Google Scholar]
- Jung T, Hansen EM, Winton L, Oßwald W, Delatour C. (2002) Three new species of Phytophthora from European oak forests. Mycological Research 106: 397–411. [Google Scholar]
- Jung T, Jung MH, Scanu B, Seress D, Kovács GM, et al. (2017) Six new Phytophthora species from ITS Clade 7a including two sexually functional heterothallic hybrid species detected in natural ecosystems in Taiwan. Persoonia 38: 100–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung T, Nechwatal J. (2008) Phytophthora gallica sp. nov., a new species from rhizosphere soil of declining oak and reed stands in France and Germany. Mycological Research 112: 1195–1205. [DOI] [PubMed] [Google Scholar]
- Jung T, Nechwatal J, Cooke DEL, Hartmann G, Blaschke M, et al. (2003) Phytophthora pseudosyringae sp. nov., a new species causing root and collar rot of deciduous tree species in Europe. Mycological Research 107: 772–789. [DOI] [PubMed] [Google Scholar]
- Jung T, Stukely MJC, Hardy GESJ, White D, Paap T, et al. (2011) Multiple new Phytophthora species from ITS Clade 6 associated with natural ecosystems in Australia: evolutionary and ecological implications. Persoonia 26: 13–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Mansfield MA, Park B, Geiser DM, Ivors KL, et al. (2010) The promise and pitfalls of sequence-based identification of plant-pathogenic fungi and oomycetes. Phytopathology 100: 732–737. [DOI] [PubMed] [Google Scholar]
- Katsura K. (1976) Two new species of Phytophthora causing damping-off of cucumber and trunk rot of chestnut. Transactions of the Mycological Society of Japan 17: 238–242. [Google Scholar]
- Kaufmann MJ, Gerdemann JW. (1958) Root and stem rot of soybean caused by Phytophthora sojae n. sp. Phytopathology 48: 201–208. [Google Scholar]
- Kennedy DM, Duncan JM. (1995) A papillate Phytophthora species with specificity to Rubus. Mycological Research 99: 57–68. [Google Scholar]
- Klebahn H. (1905) Eine neue Pilzkrankheit der Syringen (A new fungal disease of Syringae). Zentralblatt für Bakteriologie, Parasitenkunde und Infektionskrankheiten 15: 335–336. [Google Scholar]
- Ko WH, Ann PJ. (1985) Phytophthora humicola, a new species from soil of a citrus orchard in Taiwan. Mycologia 77: 631–636. [Google Scholar]
- Kröber H, Marwitz R. (1993) Phytophthora tentaculata sp. nov. und Phytophthora cinnamomi var. parvispora var. nov., zwei neue Pilze von Zierpflanzen in Deutschland. Zeitschrift Fur Pflanzenkrankheiten Und Pflanzenschutz 100: 250–258. [Google Scholar]
- Kroon LPNM, Bakker FT, Van Den Bosch GBM, Bonants PJM, Flier WG. (2004) Phylogenetic analysis of Phytophthora species based on mitochondrial and nuclear DNA sequences. Fungal Genetics and Biology 41: 766–782. [DOI] [PubMed] [Google Scholar]
- Kroon LPNM, Brouwer H, De Cock AWAM, Govers F. (2012) The genus Phytophthora anno 2012. Phytopathology 102: 348–364. [DOI] [PubMed] [Google Scholar]
- Lara E, Belbahri L. (2011) SSU rRNA reveals major trends in oomycete evolution. Fungal Diversity 49: 93–100. [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, Mcgettigan PA, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Leonian LH. (1922) Stem and fruit blight of pepper caused by Phytophthora capsici sp. nov. Phytopathology 12: 401–408. [Google Scholar]
- Li WW, Zhao WX, Huai WX. (2017) Phytophthora pseudopolonica sp. nov., a new species recovered from stream water in subtropical forests of China. International Journal of Systematic and Evolutionary Microbiology 67: 3666–3675. [DOI] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR. (2017) Mesquite: a modular system for evolutionary analysis. http://mesquiteproject.org. [Google Scholar]
- Man In’t Veld WA, De Cock A, Ilieva E, Lévesque CA. (2002) Gene flow analysis of Phytophthora porri reveals a new species: Phytophthora brassicae sp. nov. European Journal of Plant Pathology 108: 51–62. [Google Scholar]
- Man In’t Veld WA, Rosendahl KCHM, Brouwer H, De Cock AWAM. (2011) Phytophthora gemini sp. nov., a new species isolated from the halophilic plant Zostera marina in the Netherlands. Fungal Biology 115: 724–732. [DOI] [PubMed] [Google Scholar]
- Man In’t Veld WA, Rosendahl KCHM, Hong C. (2012) Phytophthora ×serendipita sp. nov. and P. ×pelgrandis, two destructive pathogens generated by natural hybridization. Mycologia 104: 1390–1396. [DOI] [PubMed] [Google Scholar]
- Man In’t Veld WA, Rosendahl KCHM, Van Rijswick PCJ, Meffert JP, Westenberg M, et al. (2015) Phytophthora terminalis sp. nov. and Phytophthora occultans sp. nov., two invasive pathogens of ornamental plants in Europe. Mycologia 107: 54–65. [DOI] [PubMed] [Google Scholar]
- Man In’t Veld WA. (2007) Gene flow analysis demonstrates that Phytophthora fragariae var. rubi constitutes a distinct species, Phytophthora rubi comb. nov. Mycologia 99: 222–226. [DOI] [PubMed] [Google Scholar]
- Martin FN, Abad ZG, Balci Y, Ivors K. (2012) Identification and detection of Phytophthora: reviewing our progress, identifying our needs. Plant Disease 96: 1080–1103. [DOI] [PubMed] [Google Scholar]
- Martin FN, Blair JE, Coffey MD. (2014) A combined mitochondrial and nuclear multilocus phylogeny of the genus Phytophthora. Fungal Genetics and Biology 66: 19–32. [DOI] [PubMed] [Google Scholar]
- Martin FN, Tooley PW. (2003) Phylogenetic relationships among Phytophthora species inferred from sequence analysis of mitochondrially encoded cytochrome oxidase I and II genes. Mycologia 95: 269–284. [PubMed] [Google Scholar]
- Maseko B, Burgess TI, Coutinho TA, Wingfield MJ. (2007) Two new Phytophthora species from South African Eucalyptus plantations. Mycological Research 111: 1321–1338. [DOI] [PubMed] [Google Scholar]
- Mcrae W. (1918) Phytophthora meadii n. sp. on Hevea brasiliensis. Memoirs of the Department of Agriculture in India, Botanical Series 9: 219–273. [Google Scholar]
- Mirabolfathy M, Cooke DEL, Duncan JM, Williams NA, Ershad D, et al. (2001) Phytophthora pistaciae sp. nov. and P. melonis: the principal causes of pistachio gummosis in Iran. Mycological Research 105: 1166–1175. [Google Scholar]
- Mostowfizadeh-Ghalamfarsa R, Cooke DEL, Banihashemi Z. (2008) Phytophthora parsiana sp. nov., a new high-temperature tolerant species. Mycological Research 112: 783–794. [DOI] [PubMed] [Google Scholar]
- Naher M, Motohash K, Watanabe H, Chikuo Y, Senda M, et al. (2011) Phytophthora chrysanthemi sp. nov., a new species causing root rot of chrysanthemum in Japan. Mycological Progress 10: 21–31. [Google Scholar]
- Nechwatal J, Bakonyi J, Cacciola SO, Cooke DEL, Jung T, et al. (2013) The morphology, behaviour and molecular phylogeny of Phytophthora taxon Salixsoil and its redesignation as Phytophthora lacustris sp. nov. Plant Pathology 62: 355–369. [Google Scholar]
- Nelson SC, Abad ZG. (2010) Phytophthora morindae, a new species causing black flag disease on noni (Morinda citrifolia L.) in Hawaii. Mycologia 102: 122–134. [DOI] [PubMed] [Google Scholar]
- Nirenberg HI, Gerlach WF, Graefenhan T. (2009) Phytophthora ×pelgrandis, a new natural hybrid pathogenic to Pelargonium grandiflorum hort. Mycologia 101: 220–231. [DOI] [PubMed] [Google Scholar]
- Oliva RF, Kroon LPNM, Chacon G, Flier WG, Ristaino JB, et al. (2010) Phytophthora andina sp. nov., a newly identified heterothallic pathogen of solanaceous hosts in the Andean highlands. Plant Pathology 59: 613–625. [Google Scholar]
- Oudemans P, Coffey MD. (1991) A revised systematics of twelve papillate Phytophthora species based on isozyme analysis. Mycological Research 95: 1025–1046. [Google Scholar]
- Pánek M, Fér T, Mráček J, Tomšovský M. (2016) Evolutionary relationships within the Phytophthora cactorum species complex in Europe. Fungal Biology 120: 836–851. [DOI] [PubMed] [Google Scholar]
- Park B, Martin F, Geiser DM, Kim HS, Mansfield MA, et al. (2013) Phytophthora database 2.0: update and future direction. Phytopathology 103: 1204–1208. [DOI] [PubMed] [Google Scholar]
- Petersen HE. (1910) An account of Danish freshwater-phycomycetes, with biological and systematical remarks. Annales Mycologici 8: 494–560. [Google Scholar]
- Pethybridge GH. (1913) On the rotting of potato tubers by a new species of Phytophthora having a method of sexual reproduction hitherto undescribed. Scientific Proceedings of the Royal Dublin Society 13: 529–565. [Google Scholar]
- Pethybridge GH, Lafferty AH. (1919) A disease of tomato and other plants caused by a new species of Phytophthora. Scientific Proceedings of the Royal Dublin Society 15: 487–505. [Google Scholar]
- Posada D. (2008) jModelTest: phylogenetic model averaging. Molecular Biology and Evolution 25: 1253–1256. [DOI] [PubMed] [Google Scholar]
- Puglisi I, De Patrizio A, Schena L, Jung T, Evoli M, et al. (2017) Two previously unknown Phytophthora species associated with brown rot of Pomelo (Citrus grandis) fruits in Vietnam. PLoS ONE 12: e0172085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purss GS. (1957) Stem rot: a disease of cowpeas caused by an undescribed species of Phytophthora. Queensland Journal of Agricultural Science 14: 125–154. [Google Scholar]
- Raciborski M. (1900) Parasitische Algen und Pilze Java’s. Buitenzorg: Botanisches Institut. [Google Scholar]
- Rahman MZ, Mukobata H, Suga H, Kageyama K. (2014a) Phytophthora asiatica sp. nov., a new species causing leaf and stem blight of kudzu in Japan. Mycological Progress 13: 759–769. [Google Scholar]
- Rahman MZ, Uematsu S, Kimishima E, Kanto T, Kusunoki M, et al. (2015) Two plant pathogenic species of Phytophthora associated with stem blight of Easter lily and crown rot of lettuce in Japan. Mycoscience 56: 419–433. [Google Scholar]
- Rahman MZ, Uematsu S, Takeuchi T, Shirai K, Ishiguro Y, et al. (2014b) Two new species, Phytophthora nagaii sp. nov. and P. fragariaefolia sp. nov., causing serious diseases on rose and strawberry plants, respectively, in Japan. Journal of General Plant Pathology 80: 348–365. [Google Scholar]
- Rands RD. (1922) Streepkanker van Kaneel, Veroorzaakt Door Phytophthora cinnamomi n. sp. Batavia: Drukkerij Ruygrok. [Google Scholar]
- Rea AJ, Burgess TI, Hardy GESJ, Stukely MJC, Jung T. (2011) Two novel and potentially endemic species of Phytophthora associated with episodic dieback of Kwongan vegetation in the south-west of Western Australia. Plant Pathology 60: 1055–1068. [Google Scholar]
- Rea AJ, Jung T, Burgess TI, Stukely MJC, Hardy GESJ. (2010) Phytophthora elongata sp. nov., a novel pathogen from the Eucalyptus marginata forest of Western Australia. Australasian Plant Pathology 39: 477–491. [Google Scholar]
- Reeser P, Sutton W, Hansen E. (2013) Phytophthora pluvialis, a new species from mixed tanoak-Douglas-fir forests of western Oregon, U.S.A. North American Fungi 8: 1–8. [Google Scholar]
- Reeser PW, Hansen EM, Sutton W. (2007) Phytophthora siskiyouensis, a new species from soil, water, myrtlewood (Umbellularia californica) and tanoak (Lithocarpus densiflorus) in southwestern Oregon. Mycologia 99: 639–643. [DOI] [PubMed] [Google Scholar]
- Rizzo DM, Garbelotto M, Davidson JM, Slaughter GW, Koike ST. (2002) Phytophthora ramorum as the cause of extensive mortality of Quercus spp. and Lithocarpus densiflorus in California. Plant Disease 86: 205–214. [DOI] [PubMed] [Google Scholar]
- Rizzo DM, Garbelotto M, Hansen EA. (2005) Phytophthora ramorum: integrative research and management of an emerging pathogen in California and Oregon forests. Annual Review of Phytopathology 43: 309–335. [DOI] [PubMed] [Google Scholar]
- Robideau GP, De Cock AWAM, Coffey MD, Voglmayr H, Brouwer H, et al. (2011) DNA barcoding of oomycetes with cytochrome c oxidase subunit I and internal transcribed spacer. Molecular Ecology Resources 11: 1002–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, et al. (2012) MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safaiefarahani B, Mostowfizadeh-Ghalamfarsa R, Hardy G, Burgess TI. (2015) Re-evaluation of the Phytophthora cryptogea species complex and the description of a new species, Phytophthora pseudocryptogea sp. nov. Mycological Progress 14: 108. [Google Scholar]
- Safaiefarahani B, Mostowfizadeh-Ghalamfarsa R, Hardy GES, Burgess TI. (2016) Species from within the Phytophthora cryptogea complex and related species, P. erythroseptica and P. sansomeana, readily hybridize. Fungal Biology 120: 975–987. [DOI] [PubMed] [Google Scholar]
- Sawada K. (1927) Descriptive catalogue of the Formosan fungi III. Report of the Department of Agriculture Government Research Institute of Formosa 27: 1–62. [Google Scholar]
- Scanu B, Hunter GC, Linaldeddu BT, Franceschini A, Maddau L, et al. (2014) A taxonomic re-evaluation reveals that Phytophthora cinnamomi and P. cinnamomi var. parvispora are separate species. Forest Pathology 44: 1–20. [Google Scholar]
- Scanu B, Linaldeddu BT, Deidda A, Jung T. (2015) Diversity of Phytophthora species from declining Mediterranean maquis vegetation, including two new species, Phytophthora crassamura and P. ornamentata sp. nov. PLoS ONE 10: e0143234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröter J. (1886) Die Pilze Schlesiens In: Kryptogamen-Flora von Schlesien (Cohn F, ed.): 3 (1): 1–814. Breslau: J U Kern’s Verlag. [Google Scholar]
- Scott PM, Burgess TI, Barber PA, Shearer BL, Stukely MJC, et al. (2009) Phytophthora multivora sp. nov., a new species recovered from declining Eucalyptus, Banksia, Agonis and other plant species in Western Australia. Persoonia 22: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simamora AV, Stukely MJC, Hardy GES, Burgess TI. (2015) Phytophthora boodjera sp. nov., a damping-off pathogen in production nurseries and from urban and natural landscapes, with an update on the status of P. alticola. IMA Fungus 6: 319–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RE, Smith EH. (1906) A new fungus of economic importance. Botanical Gazette 42: 215–221. [Google Scholar]
- Stamatakis A. (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. (2002) PAUP*: phylogenetic analysis using parsimony (*and other methods). Sunderland, MA: Sinauer Associates. [Google Scholar]
- Taylor PA, Pascoe IG, Greenhalgh FC. (1985) Phytophthora clandestina sp. nov. in roots of subterranean clover. Mycotaxon 22: 77–85. [Google Scholar]
- Thaxter R. (1889) A new American Phytophthora. Botanical Gazette 14: 273–274. [Google Scholar]
- Thompson A. (1929) Phytophthora species in Malaya. The Malayan Agricultural Journal 17: 53–100. [Google Scholar]
- Tomlinson JA. (1952) Brown core root rot of Primula caused by Phytophthora primulae n. sp. Transactions of the British Mycological Society 35: 221–235. [Google Scholar]
- Tucker CM. (1931) Taxonomy of the Genus Phytopthora de Bary. Research Bulletin of the Missouri Agricultural Experiment Station 153: 1–208. [Google Scholar]
- Tucker CM, Milbrath JA. (1942) Root rot of Chamaecyparis caused by a species of Phytophthora. Mycologia 34: 94–103. [Google Scholar]
- Turner RS. (2005) After the famine: Plant pathology, Phytophthora infestans, and the late blight of potatoes, 1845–1960. Historical Studies in the Physical and Biological Sciences 35: 341–370. [Google Scholar]
- Tyler BM, Tripathy S, Zhang XM, Dehal P, Jiang RHY, et al. (2006) Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 313: 1261–1266. [DOI] [PubMed] [Google Scholar]
- Uzuhashi S, Tojo M, Kakishima M. (2010) Phylogeny of the genus Pythium and description of new genera. Mycoscience 51: 337–365. [Google Scholar]
- Vettraino AM, Brasier CM, Brown AV, Vannini A. (2011) Phytophthora himalsilva sp. nov. an unusually phenotypically variable species from a remote forest in Nepal. Fungal Biology 115: 275–287. [DOI] [PubMed] [Google Scholar]
- Villa NO, Kageyama K, Asano T, Suga H. (2006) Phylogenetic relationships of Pythium and Phytophthora species based on ITS rDNA, cytochrome oxidase II and beta-tubulin gene sequences. Mycologia 98: 410–422. [DOI] [PubMed] [Google Scholar]
- Waterhouse GM. (1963) Key to the species of Phytophthora de Bary. Mycological Papers 92: 1–22. [Google Scholar]
- Weir BS, Paderes EP, Anand N, Uchida JY, Pennycook SR, et al. (2015) A taxonomic revision of Phytophthora Clade 5 including two new species, Phytophthora agathidicida and P. cocois. Phytotaxa 205: 21–38. [Google Scholar]
- Werres S, Marwitz R, Man in’t Veld WA, De Cock AW, Bonants PJ, et al. (2001) Phytophthora ramorum sp. nov., a new pathogen on Rhododendron and Viburnum. Mycological Research 105: 1155–1165. [Google Scholar]
- Yang X. (2014) New Species and Phylogeny of the Genus Phytophthora. PhD thesis, Virginia Tech. [Google Scholar]
- Yang X, Balci Y, Brazee NJ, Loyd AL, Hong C. (2016) A unique species in Phytophthora clade 10, Phytophthora intercalaris sp. nov., recovered from stream and irrigation water in the eastern USA. International Journal of Systematic and Evolutionary Microbiology 66: 845–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Copes WE, Hong CX. (2013) Phytophthora mississippiae sp. nov., a new species recovered from irrigation reservoirs at a plant nursery in Mississippi. Journal of Plant Pathology & Microbiology 4: 180. [Google Scholar]
- Yang X, Copes WE, Hong CX. (2014a) Two novel species representing a new clade and cluster of Phytophthora. Fungal Biology 118: 72–82. [DOI] [PubMed] [Google Scholar]
- Yang X, Gallegly ME, Hong CX. (2014b) A high-temperature tolerant species in clade 9 of the genus Phytophthora: P. hydrogena sp. nov. Mycologia 106: 57–65. [DOI] [PubMed] [Google Scholar]
- Yang X, Hong C. (2014) Halophytophthora fluviatilis sp. nov. from freshwater in Virginia. FEMS Microbiology Letters 352: 230–237. [DOI] [PubMed] [Google Scholar]
- Yang X, Hong CX. (2013) Phytophthora virginiana sp. nov., a high-temperature tolerant species from irrigation water in Virginia. Mycotaxon 126: 167–176. [Google Scholar]
- Yang X, Richardson PA, Hong C. (2014c) Phytophthora ×stagnum nothosp. nov., a new hybrid from irrigation reservoirs at ornamental plant nurseries in Virginia. PLoS ONE 9: e103450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye W, Wang Y, Shen D, Li D, Pu T, et al. (2016) Sequencing of the litchi downy blight pathogen reveals it is a Phytophthora species with downy mildew-like characteristics. Molecular Plant-Microbe Interactions 29: 573–583. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


