Fig. 3.
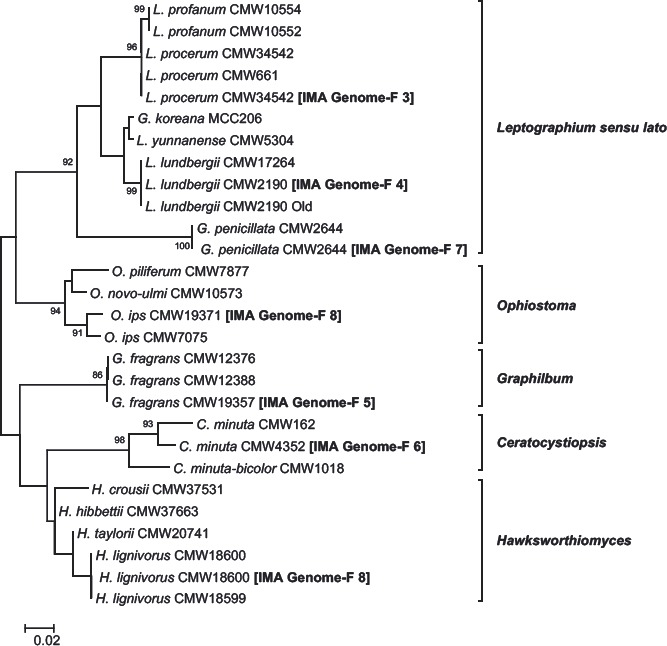
Identity verification of Ophiostomataceae isolates sequenced in this study and in all previous IMA Genome Announcements (IMA Genome-F: 3–7; Van der Nest et al. 2014a, Wingfield et al. 2015a, 2016a, 2015b, 2016b). Gene regions (LSU, βT) used for verification were extracted from assembled genomes. Other reference isolates and their corresponding sequences were obtained from published papers (De Beer et al. 2016, Linnakoski et al. 2012, Yin et al. 2015, Zipfel et al. 2006). The phylogeny was constructed using RAxML with the GTRGAMMA substitution model. Bootstrap values greater than 75 are indicated at the nodes.
