Abstract
Objectives
The role of long non-coding RNA (lncRNA) expression in human head and neck squamous cell carcinoma (HNSCC) is still poorly understood. In this study, we aimed at establishing the onco-lncRNAome profiling of HNSCC and to identify lncRNAs correlating with prognosis and patient survival.
Materials and Methods
The Atlas of Noncoding RNAs in Cancer (TANRIC) database was employed to retrieve the lncRNA expression information generated from The Cancer Genome Atlas (TCGA) HNSCC RNA-sequencing data. RNA-sequencing data from HNSCC cell lines were also considered for this study. Bioinformatics approaches, such as differential gene expression analysis, survival analysis, principal component analysis, and Co-LncRNA enrichment analysis were performed.
Results
Using TCGA HNSCC RNA-sequencing data from 426 HNSCC and 42 adjacent normal tissues, we found 728 lncRNA transcripts significantly and differentially expressed in HNSCC. Among the 728 lncRNAs, 55 lncRNAs were significantly associated with poor prognosis, such as overall survival and/or disease-free survival. Next, we found 140 lncRNA transcripts significantly and differentially expressed between Human Papilloma Virus (HPV) positive tumors and HPV negative tumors. Thirty lncRNA transcripts were differentially expressed between TP53 mutated and TP53 wild type tumors. Co-LncRNA analysis suggested that protein-coding genes that are co-expressed with these deregulated lncRNAs might be involved in cancer associated molecular events. With consideration of differential expression of lncRNAs in a HNSCC cell lines panel (n=22), we found several lncRNAs that may represent potential targets for diagnosis, therapy and prevention of HNSCC.
Conclusion
LncRNAs profiling could provide novel insights into the potential mechanisms of HNSCC oncogenesis.
Keywords: Long non-coding RNA, RNA-sequencing, head and neck squamous cell carcinoma (HNSCC), human papilloma virus (HPV), TP53, TCGA
INTRODUCTION
More than 59,000 new cases of head and neck cancers are estimated for 2015 in the US alone, including 45,000 cases arising in the oral cavity and pharynx and 13,000 in the larynx, resulting each year in approximately 11,000 cancer-associated deaths (http://seer.cancer.gov/). Most head and neck malignancies are squamous cell carcinoma (HNSCC), which remain associated with a poor outcome [1], primarily due to local tumor recurrence and regional lymph node and distant metastasis, despite of considerable advances in multimodality therapy including surgery, radiation therapy and chemotherapy to control local disease [2]. Therefore, understanding the molecular mechanisms underlying HNSCC could help to improve diagnosis, and facilitate the development of new strategies to treat and prevent this disease.
The non-coding regions of the human genome were previously considered as the “junk” or “noise” [3]. However, along with the advance of comprehensive sequencing and high resolution microarray technologies, it has been demonstrated that 98% of the human genome is part of the non-coding region [4]. Approximately 70% of the genome is actively transcribed according to the ENCODE project [5]. The proportion of non-coding sequences that are actively transcribed increases with eukaryotes complexity [6]. Therefore, understanding the non-coding RNAs (ncRNAs) world has become essential. NcRNAs are classified broadly into two groups based on the size of the transcript: short ncRNAs (<200nt) and long ncRNAs (lncRNAs) (>200nt). The group of short ncRNA including microRNA (miRNA), piwi-interacting RNA (piRNA), and small interfering RNA (siRNA) has attracted considerable attention in the last few years, as miRNAs, for example, play important regulatory roles in cancer and other diseases [7-9].
LncRNAs are generally defined as endogenous non-coding RNA molecules longer than 200nt in length. To date 14,880 lncRNAs from 9,277 loci have been annotated by the GENCODE project [10]. Most of lncRNAs are transcribed by RNA polymerase II, then polyadenylated and pre-RNA spliced. The majority of lncRNAs are located in cell nucleus, but some of lncRNAs are in both nucleus and cytoplasm, and some lncRNAs locate to the cytoplasm specifically. Their expression patterns are highly tissue-specific [11], with lncRNA expression generally lower than that for mRNA [12].
LncRNAs are classified based on their genomic position related with neighboring genes (e.g. intergenic, exonic, intronic or overlapping genes) and also based on their structural or functional features (e.g., circular RNAs, antisense RNAs, transcribed ultraconserved noncoding RNAs (T-UCRs), long enhancer ncRNAs, long intergenic ncRNAs (lincRNAs) and pseudogenes) [10,13]. LncRNAs are involved in diverse biological processes [14]. They can negatively or positively affect expression of protein-coding gene by transcriptional interference or by chromatin modification. For example, antisense transcripts can hybridize to their specific pre-mRNAs resulting in alternatively spliced mRNAs or generating endogenous siRNAs through Dicer [15]. LncRNAs can silence miRNA expression as “miRNA sponges” [16]. They can also modulate protein activity and cellular localization [14]. Moreover, lncRNAs can be processed to small ncRNAs that may act as endo-siRNAs or miRNAs [17]. Ultimately, lncRNAs are involved in multiple structural and/or organizational roles in the cell, and emerging reports indicate that lncRNAs are dysregulated in various diseases [18,19]. Although the complexity of the biological functions of lncRNAs are still mostly unknown, accumulating studies suggest that lncRNAs contribute to the initiation and development of various cancer types by acting as oncogenic or tumor suppressive RNAs [20-24].
While in the past genetic analyses in HNSCC had been often performed in small cohorts of patients in single platforms, the recent availability of large publically available cancer data sets enabled by the TCGA (The Cancer Genome Atlas), has revolutionized cancer biology as we know it [25]. TCGA was launched in 2006, as part of a coordinated effort of the National Cancer Institute (NCI) and the National Human Genome Research Institute (NHGRI), both part of the National Institutes of Health (NIH) in the United States (http://cancergenome.nih.gov/abouttcga). TCGA is a comprehensive project aimed at increasing the understanding of the molecular mechanism of cancer through the use of multiple platform technologies, including large-scale genome sequencing, in clinically annotated cancer specimens. This integrated genomic analysis has accelerated the discovery and validation of novel molecular mechanisms driving cancer progression in multiple cancer types. In this context, the TCGA project team has recently published the comprehensive genomic analysis of HNSCC from 279 patients [26].
In this study, we investigated deregulated expression of lncRNAs during HNSCC progression by analyzing the emerging sequence information from normal and HNSCC cancer lesions in the TCGA databases. This HNSCC lncRNAome profiling revealed remarkable tissue specificity of lncRNA expression in both normal and malignant tissues, and that HPV status and genomic alterations affect the lncRNA expression patterns. These results might provide a new avenue for diagnosis and therapeutic intervention in HNSCC.
MATERIALS AND METHODS
Data retrieval of lncRNA profiles from HNSCC cancer cell lines and clinical samples
We used The Atlas of Noncoding RNAs in Cancer (TANRIC) [27] to retrieve the lncRNA expression information from the TCGA HNSCC RNA-sequencing database. After retrieving the data, we annotated each of the samples according to their barcode ID based on the available clinical information, including overall survival (OS), disease-free survival (DFS) status, clinical stage, the HPV infection status, and the presence of gene mutations in HNSCC-relevant genes, such as TP53, and PIK3CA from cBioPortal [28]. Patient and tissue sample characteristics are described in Supplementary Table 1. The procedure of RNA-sequencing profiling from a large panel of HNSCC-derived cells from different anatomical locations and human papillomavirus (HPV) infection status (herein referred as oral and pharyngeal cancer; OPC-22 panel) was previously described [29].
Statistical and data mining analyses of lncRNA profiles among HNSCC
We utilized the SAM test to identify differentially expressed lncRNAs among normal and HNSCC and additional factors such as: HPV infection and TP53 mutation. Statistical analysis and heatmap visualization of differentially expressed transcripts were done with the MultiExperiment Viewer software (MeV 4.9) [30].
To further explore the prognostic value of the lncRNAs in HNSCC, a group of 418 patients with HNSCC for OS or 301 out of 418 patients with HNSCC for DFS with follow-up data were divided in two subgroups according to the median expressions of each lncRNA analyzed. These groups were then analyzed by univariate (Kaplan–Meier survival curves and log-rank statistics) and multivariate (Cox proportional hazards) methods using the Survival R package. The multivariate Cox proportional-hazard model included: gender, anatomic location, tumor pathological stage, TP53 mutation status and gene expression of the lncRNA under consideration. In this model, HPV infection was not included because of the limited number of samples with HPV infection data. Overall survival and disease-free survival were the end points.
The Co-LncRNA online resource was employed to identify bioprocess modulated by lncRNAs. Briefly, Co-LncRNA is a web-based tool that performs Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses based on co-expressed protein-coding genes (CEGs) of a single or multiple lncRNAs [31]. The REViGO resource was employed to summarize and visualize the enriched GO terms in a scatterplot graph based on the corrected p-values obtained by Co-LncRNA resource [32].
RESULTS
Differential expression and prognosis associated lncRNAs in Tumor vs. Normal analysis
First, we employed the data from 468 HNSCC samples in TCGA RNA-sequencing data generated by Illumina HiSeq 2000. Statistical analysis of lncRNA profiles revealed 728 transcripts differentially expressed between normal and tumor samples (212 up-modulated and 516 down-modulated lncRNAs). The most statistically significant transcripts deregulated between normal and tumor samples are represented in Figure 1B, and all relevant information can be found in Supplementary Table 2 (using a cutoff of absolute fold-change > 1.5 and q-value < 0.001). Sub-organ specifically expressed lncRNAs are also shown in Figure 1A and described in Supplementary table 2. GO enrichment analysis for protein-coding genes that are co-expressed with the deregulated lncRNAs was performed by the Co-LncRNA algorithm [31], which identifies CEGs that are significantly related to gene expression, RNA splicing, protein metabolism, Tie/Notch/Wnt pathways, apoptosis, cell cycle, endocytosis, and trafficking activities, which were summarized and visualized by REViGO approach (Figure 1C and Supplementary table 2).
Figure 1.
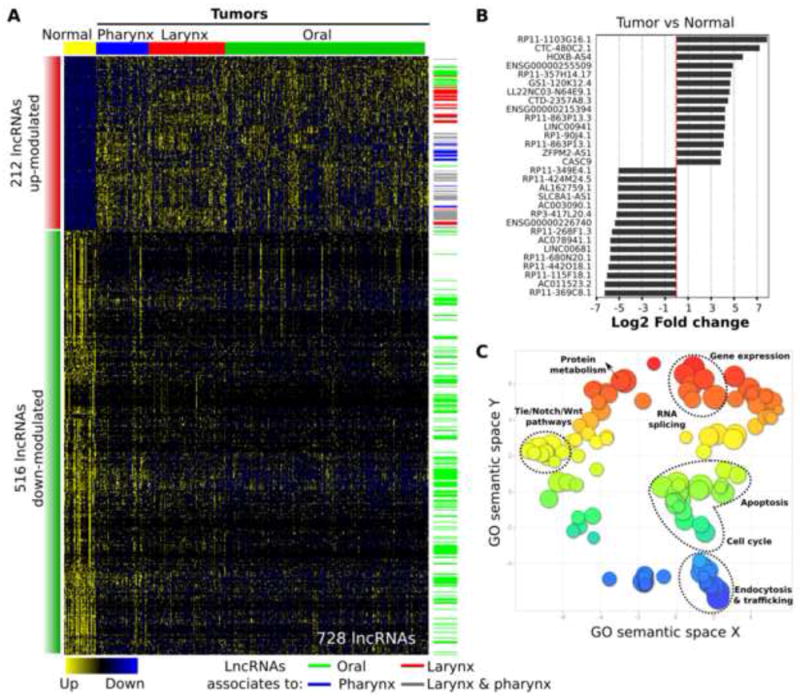
Differential expression analysis between tumor and normal tissues in the HNSCC TCGA.
(A) Heatmap of 728 differentially expressed lncRNAs in comparison between tumor and normal tissues. (B) Representative differentially expressed lncRNAs between tumor and normal tissues. (C) Gene Ontology (GO) enrichment analysis for protein-coding genes co-expressed with 728 lncRNAs with visualization by REViGO algorism.
We found 55 lncRNAs which have significant relationship with patient’s prognosis in OS or DFS (log-rank test p-value < 0.05) among 728 deregulated lncRNAs (Supplementary Table 2). To further evaluate the independent prognostic value of these 55 lncRNAs, we next performed a multivariate Cox proportional-hazard analysis that included the traditional prognostic factors such as: tumor pathological stage, anatomic location, TP53 mutation status and gender. This analysis demonstrated that 23 out of 55 lncRNAs were independent predictors of overall survival (Supplementary Table 2).
Representative genes are listed in Table 1. Kaplan-Meier plots of representative prognosis-associated lncRNAs are shown in Figure 2A. KEGG pathway enrichment analysis based on Co-LncRNA algorism for the protein-coding genes which are co-expressed with the prognosis associated lncRNAs suggests that those CEGs are significantly linked to several oncogenic events, such as cell cycle, spliceosome, endocytosis, regulation of actin cytoskeleton, MAPK, Wnt, VEGF, Neutrophin, Insulin, ERBB, and Notch signaling (Figure 2B). Among these 728 lncRNAs, eight lncRNA genes (ADAMTS9-AS2, CBR3-AS1, DLEU2, FGF10-AS1, HCP5, LINC00271, PDZRN3-AS1, RMST, TUG1, TUSC8) were matched with the list of LncRNADisease database [33], which are LncRNAs previously reported in relationship with disease including several malignancies (Supplementary Table 3). We briefly summarize the current knowledge of these genes in the discussion section.
Table 1.
Representative lncRNAs associated with poor prognosis among 55 lncRNAs.
| ENSEMBL ID | Gene name | Fold change (Tumor/Normal) | Survival outcome | Log-rank p-value | Cox p-value | HR (95% CI) |
|---|---|---|---|---|---|---|
| LncRNAs up-modulated in tumors | ||||||
| ENSG00000250874 | CTC-480C2.1 | 148.6 | Reduced OS | 0.0006 | 0.1447 | --- |
| ENSG00000271127 | LL22NC03-N64E9.1 | 24.4 | Reduced OS | 0.0247 | 0.0351 | 1.5 (1.03 – 2.27) |
| ENSG00000261327 | RP11-863P13.3 | 18.5 | Reduced DFS | 0.0423 | 0.133 | --- |
| ENSG00000260162 | RP11-863P13.1 | 17.1 | Reduced DFS | 0.015 | 0.0576 | --- |
| ENSG00000249395 | CASC9 | 14.5 | Reduced OS | 0.0047 | 0.0498 | 1.5 (1 – 2.18) |
| ENSG00000256268 | RP11-221N13.3 | 14.1 | Reduced OS | 0.0462 | 0.686 | --- |
| ENSG00000267284 | RP11-397A16.1 | 12.4 | Reduced OS | 0.0109 | 0.0457 | 1.5 (1 – 2.23) |
| ENSG00000250546 | RP11-8L2.1 | 11.2 | Reduced OS | 0.0213 | 0.1138 | --- |
| ENSG00000234695 | AC002076.10 | 9.2 | Reduced OS | 0.0204 | 0.0285 | 1.5 (1.05 – 2.30) |
| ENSG00000225548 | AC098973.2 | 8.8 | Reduced OS | 0.016 | 0.2062 | --- |
| ENSG00000234902 | AC007879.2 | 5.7 | Reduced OS | 0.0009 | 0.112 | --- |
| ENSG00000271826 | PLS3-AS1 | 4.2 | Reduced OS | 0.019 | 0.5359 | --- |
| ENSG00000251381 | LINC00958 | 3.5 | Reduced OS | 0.0139 | 0.3 | --- |
| ENSG00000237152 | DLEU7-AS1 | 3 | Reduced OS | 0.0018 | 0.00348 | 1.8 (1.2 – 2.6) |
| ENSG00000230002 | ALMS1-IT1 | 2.9 | Reduced OS | 0.0107 | 0.0102 | 1.7 (1.13 – 2.51) |
| ENSG00000245522 | RP11-540A21.2 | 2.9 | Reduced OS | 0.0408 | 0.4654 | --- |
| ENSG00000232725 | U52111.14 | 2.8 | Reduced DFS | 0.0432 | 0.21 | --- |
| ENSG00000254551 | RP11-727A23.7 | 2.7 | Reduced DFS | 0.0203 | 0.306 | --- |
| ENSG00000259901 | --- | 2.6 | Reduced OS & DFS | <0.05 | 0.089 (OS) 0.542 (DFS) | --- |
| ENSG00000172965 | MIR4435-1HG | 2.2 | Reduced OS & DFS | <0.003 | 0.0955 | --- |
|
| ||||||
| LncRNAs down-modulated in tumors | ||||||
| ENSG00000258616 | RP11-369C8.1 | -76.1 | Increased DFS | 0.0371 | 0.297 | --- |
| ENSG00000228789 | HCG22 | -21.9 | Increased OS | 0.002 | 0.2799 | --- |
| ENSG00000233987 | AC106706.1 | -21.1 | Increased OS | 0.0237 | 0.01534 | 0.41 (0.19 – 0.84) |
| ENSG00000231062 | AC103563.9 | -15.5 | Increased OS | 0.0127 | 0.0905 | --- |
| ENSG00000253389 | RP11-930P14.1 | -12.7 | Increased OS | 0.0388 | 0.09896 | --- |
| ENSG00000225129 | RP4-614C15.2 | -9.1 | Increased OS | 0.0101 | 0.0122 | 0.31 (0.12 – 0.77) |
| ENSG00000237530 | RP3-449H6.1 | -8.7 | Increased OS | 0.0093 | 0.05069 | --- |
| ENSG00000197585 | AC107218.3 | -8.1 | Increased OS | 0.0234 | 0.0238 | 0.62 (0.41 – 0.94) |
| ENSG00000259459 | RP11-321G12.1 | -8 | Increased OS | 0.0347 | 0.15003 | --- |
| ENSG00000260402 | RP11-56L13.1 | -7.4 | Increased OS | 0.0117 | 0.00206 | 0.54 (0.37 – 0.80) |
| ENSG00000223678 | RP11-311H10.4 | -6.8 | Increased OS | 0.0382 | 0.00528 | 0.45 (0.26 – 0.79) |
| ENSG00000266114 | RP11-963H4.5 | -6.5 | Increased OS | 0.0466 | 0.042 | 0.66 (0.45 – 0.98) |
| ENSG00000248799 | CTC-548H10.2 | -5.7 | Increased OS & DFS | <0.05 | 0.8832 (OS) 0.182 (DFS) | --- |
| ENSG00000249797 | CTD-3179P9.1 | -5.5 | Increased OS | 0.0059 | 0.00816 | 0.59 (0.39 – 0.87) |
| ENSG00000262133 | RP11-676J12.6 | -5 | Increased OS | 0.0343 | 0.916 | --- |
| ENSG00000232803 | SLCO4A1-AS1 | -4.4 | Increased DFS | 0.0019 | 0.145 | --- |
| ENSG00000233070 | ZFY-AS1 | -4.3 | Increased OS | 0.0004 | 0.00389 | 0.50 (0.32 – 0.80) |
| ENSG00000233529 | HCG21 | -3.4 | Increased OS | 0.0108 | 0.1142 | --- |
| ENSG00000260931 | --- | -3.4 | Increased OS | 0.0035 | 0.01234 | 0.61 (0.41 – 0.90) |
| ENSG00000230790 | AC012456.4 | -3.3 | Increased OS & DFS | <0.02 | 0.10733 (OS) 0.796 (DFS) | --- |
CI: Confidence interval
DFS: Disease-free survival
HR: Hazard ratio
OS: Overall survival
Figure 2.
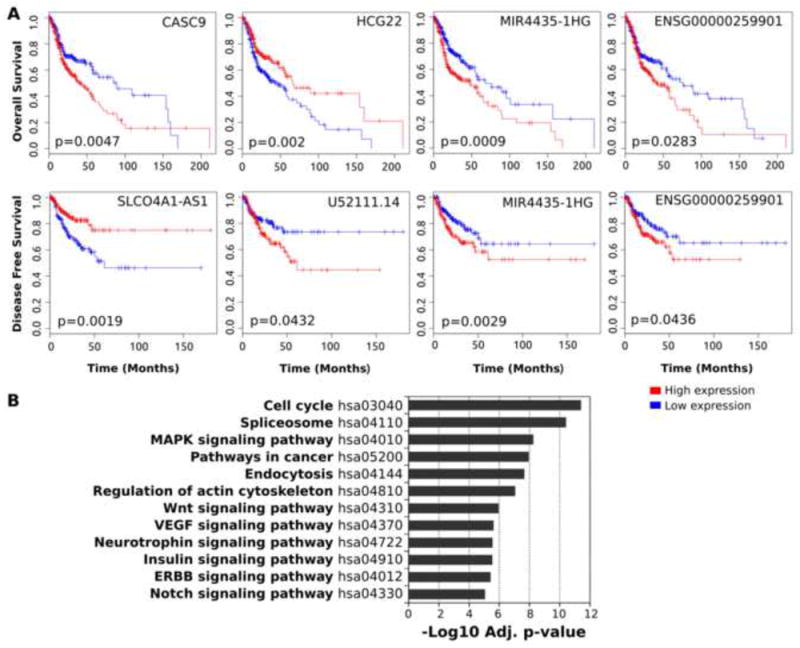
Overall survival (OS) and disease-free survival (DFS) plots and functional annotation of co-expressed protein-coding genes which are neighbor of 55 prognosis-associated lncRNAs.
(A) Representative Kaplan-Meier plots were shown in OS (upper row), and DFS (lower row). (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis is shown in bar chart for protein-coding genes co-expressed with 55 prognosis-associated lncRNAs.
Differential expression in HNSCC cell lines and HPV-specific lncRNAs
Recently, we have performed the full exome and transcriptome sequencing of OPC-22 panel [29]. Principal component analysis (PCA) showed that the cluster of NOKSI (normal oral keratinocyte spontaneously immortalized) as a control was clearly different from that of HNSCC cell lines (Figure 3A). Aberrantly expressed 30 lncRNAs in both HNSCC cell lines and tumor tissues in TCGA were shown as colored dots in volcano plots by p < 0.05 (Figure 3B and Supplementary Table 4). The role of HPV on lncRNA expression in HNSCC is largely unknown. Hence, we used the opportunity to analyze the differential expression profile between HPV positive (HPV+) and HPV negative (HPV-) HNSCC cells in the OPC-22 panel, and HPV+ and HPV- HNSCC tumors in TCGA. In the OPC-22 panel, PCA shows distinct position of component among control, HPV+, and HPV- cells (Figure 3A). There were 27 upregulated lncRNAs in HPV+ cells by absolute fold-change > 1.5, and q < 0.001 (Figure 3C). Among them, LINC01305, LINC01089, and PTOV1-AS1 were also significantly up-modulated in HPV+ tumor samples in TCGA. Overall, there were 140 up-modulated lncRNAs in HPV+ tumors in TCGA by absolute fold-change > 1.5, and q < 0.001 (Figure 3D, and supplementary table 5).
Figure 3.
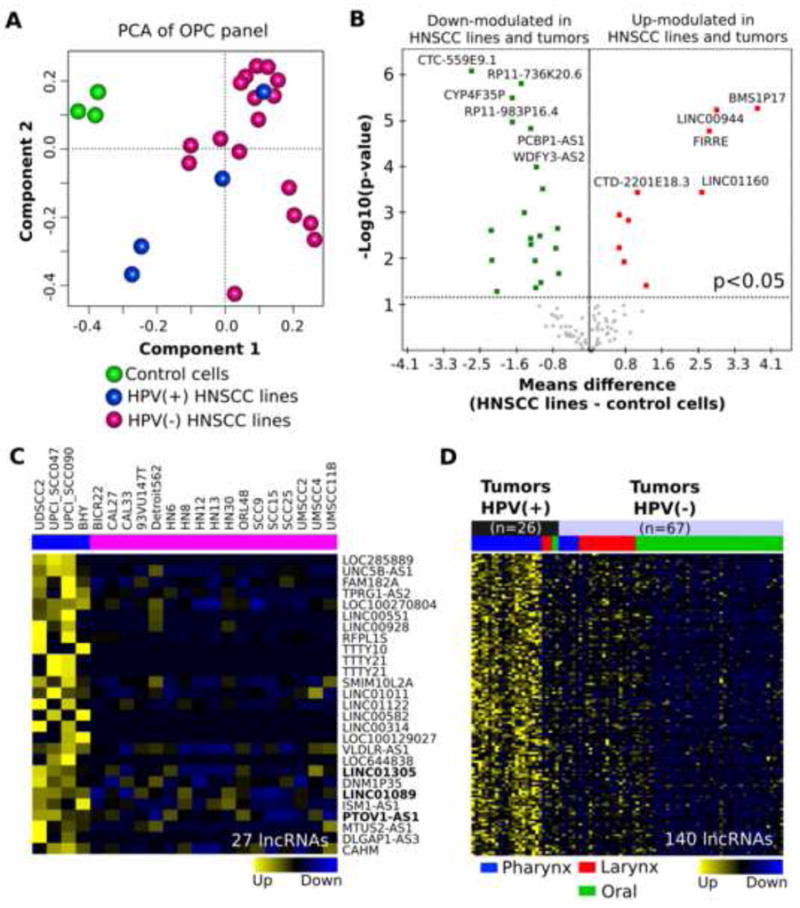
Differential expression analysis between HPV status in the OPC-22 panel and the TCGA HNSCC.
(A) Principal component analysis (PCA) among NOKSI as control cells (green), HPV+ cells (blue) and HPV- cells (pink). (B) Volcano plots shows the means difference of expression in HNSCC cells vs. control cells in X axis, and significance in HNSCC cells vs. control cells in Y axis. Broken line indicates p-value < 0.05. (C) Heatmap of 27 differentially expressed lncRNAs in comparison between HPV+ and HPV- cells. LncRNAs in bold are commonly up-modulated in HPV+ cells and tumors in TCGA HNSCC. (D) Heatmap of 140 differentially expressed lncRNAs in comparison between HPV+ and HPV- tumors in TCGA.
Differential lncRNA expression depending on TP53 gene mutation status in HNSCC
Alterations in TP53 gene are particularly of relevance in HNSCC with regards to tobacco carcinogens and HPV-infection [34]. Most HNSCC cases associated with tobacco use harbor TP53 mutations, while most HPV+ HNSCC exhibit wild type TP53. There were 30 down-modulated lncRNAs in TP53 mutated (non-silent mutation curated by Broad Institute) tumors compared with TP53 wild type tumors by absolute fold-change > 1.5, and q < 0.001 (Figure 4A, 4C and Supplementary table 6). Among the 30 lncRNAs, 19 transcripts are associated with HPV+ tumors (Figure 4B). KEGG pathway enrichment analysis based on Co-LncRNA algorism for the protein-coding genes which are co-expressed with the TP53 mutation-associated lncRNAs surprisingly indicated that these protein-coding genes were significantly related to p53-regulated signaling pathways (Figure 4D).
Figure 4.
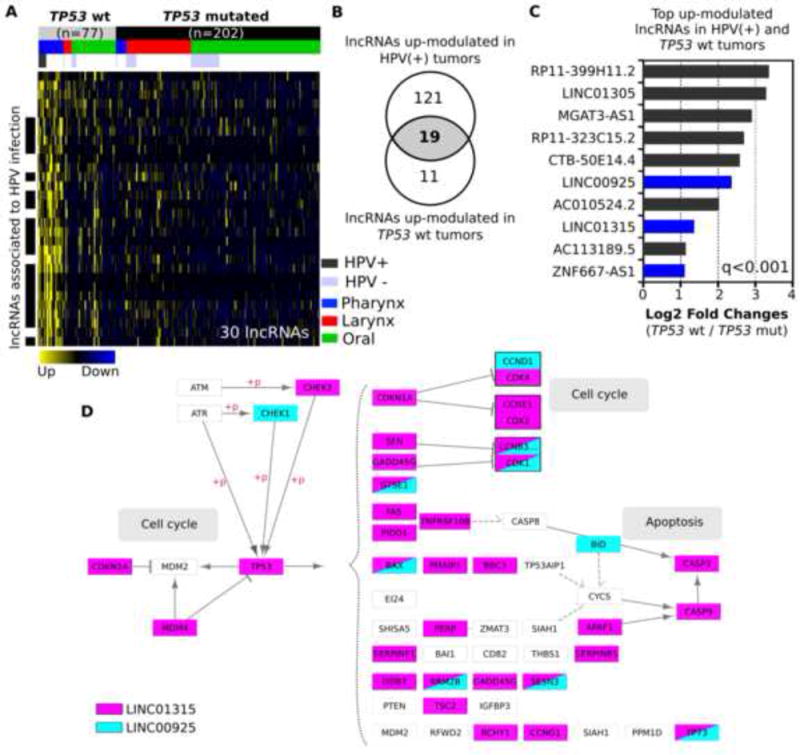
Differential expression analysis based on TP53 mutation status in the HNSCC TCGA.
(A) Heatmap of 42 differentially expressed lncRNAs in comparison between TP53 mutation and TP53 wild type. Sub organ site information (Pharynx: blue, Larynx: red, and Oral: green) and HPV infection status information (HPV+: dark gray, and HPV-: light blue) are included. (B) Venn diagram shows 19 overlapping up-modulated lncRNAs both in HPV+ and TP53 wild type tumors. (C) Representative up-modulated lncRNAs in TP53 wild type tumors are shown in bar chart. LncRNAs whose bars are in blue are significantly enriched in KEGG-P53 signaling pathway. (D) Co-expressed genes with LINC01315 or LINC00925 in KEGG-P53 signaling pathway (LINC01315: pink, and LINC00925: blue).
DISCUSSION
Recently, a growing body of evidence indicates that lncRNAs are associated with diverse and essential functions in cell biology [35]. Previous studies on lncRNAs in HNSCC have been focused on known cancer-related transcripts. For example, HOTAIR was found to be upregulated in laryngeal squamous cell carcinoma (SCC) when compared to adjacent normal tissues. There was a significant association between high levels of HOTAIR and poor prognosis in laryngeal SCC patients. Furthermore, HOTAIR regulates DNA methylation at the PTEN promoter, which leads to decreased expression of the tumor suppressor, PTEN [36]. Similarly, UCA-1, MALAT-1, NEAT-1, MEG3 [37-39], and GAS5 [40] were previously reported as dysregulated in HNSCC. In this regard, to further clarify the global expression of lncRNAs in this cancer type, we investigated the lncRNA expression signatures in the emerging TCGA HNSCC RNA-sequencing information [27] and in a wide variety of HNSCC cell lines [29]. Of interest, we failed to identify those previously mentioned transcripts as differentially expressed across our large (more than 400 cases) cohort, which was similar to the recent study in which a comprehensive analysis of lncRNAs expression was conducted in multiple HNSCC cases [41]. Thus, some lncRNA subsets previously identified in small sample collections are likely of direct relevance to the particular local risk factors or genetic variations, but not to the general aspects of HNSCC pathophysiology in the overall patient population. Here, we successfully found a number of novel lncRNAs which were associated with HNSCC oncogenesis and progression.
Overall, we found 516 upregulated and 212 downregulated lncRNAs in HNSCC samples based on 12,727 lncRNAs already registered in the TANRIC database [27]. Among the 728 lncRNAs, some lncRNAs have been documented to be associated with human diseases, including malignancies (Supplementary table 3). For example, TUG1 acts as an oncogene through promoting cell proliferation of esophageal SCC [42], bladder cancer [43], hepatocellular carcinoma (HCC) [44], and osteosarcoma [45]. TUG1 expression has been reported to be regulated by p53 in non-small cell lung carcinoma (NSCLC) [46] and SP1 transcription factor in HCC [44]. CBR3-AS1 (also known as PlncRNA-1) expression was significantly higher in prostate cancer cells relative to normal prostate epithelial cells, as well as higher in human prostate cancer tissues compared with normal tissues and benign prostatic hyperplasia [47]. The expression of CBR3-AS1 was also upregulated in human esophageal SCC compared with the adjacent noncancerous tissues [48]. However, in our current result, CBR3-AS1 was significantly downregulated in HNSCC tumor tissues. Probably, those differences might be due to a cancer type specific expression. ADAMTS9-AS2, which was down-modulated in HNSCC tumors, was reported as a new tumor suppressive lncRNA regulated by DNA-Methyltransferase 1 (DNMT1) in glioma [49]. TUSC8 (XLOC_010588) expression was significantly downregulated in cervical cancer [50], and it was consistently down-modulated in HNSCC tumors.
We identified 55 prognosis-associated lncRNAs in the HNSCC TCGA, and among them, the expression of 23 transcripts were independently associated with overall risk of death in HNSCC patients regardless of gender, organ site, tumor pathological stage, and TP53 gene mutation status. Our enrichment analysis indicates that those lncRNAs are co-expressed with protein-coding genes involved in several oncogenic signaling pathways. This suggests that these lncRNAs might play important and functional roles by regulating directly or indirectly key protein-coding gene expression that in turn may contribute to HNSCC. For example, CASC9 (ESCCAL-1), which we identified in our analysis of HNSCC, was also overexpressed in esophageal SCC, where it functions as an onco-lncRNA through inhibition of apoptosis and promoting invasion in vitro [51].
We also focused on the common deregulated lncRNAs in both HNSCC TCGA and OPC-22 panel. Among the thirty aberrantly expressed transcripts, FIRRE (Functional Intergenic Repeating RNA Element) was found to be up-modulated in both tumor samples and HNSCC cell lines. Recently, FIRRE was reported to interact with the nuclear matrix factor hnRNPU [52]. FIRRE localizes across at least three distinct trans-chromosomal loci. Both genomic excision of FIRRE locus and knockdown of hnRNPU shows decreased co-localization of these trans-chromosomal interacting loci [52]. Thus, aberrant epigenetic alterations caused by deregulated FIRRE levels may represent a novel HNSCC oncogenic mechanism, which should be clarified in future experimental studies.
The role of HPV infection on lncRNA expression in HNSCC remains unclear. To the best of our knowledge, our analysis provides the first evidence of the existence of a differential expression profile based on the HPV infection status using both HNSCC cell lines (OPC-22 panel) and clinical samples from the TCGA HNSCC. HPV+ tumors were predominantly from pharyngeal site (80.8%) and did not exhibit mutated TP53, whereas HPV- tumors were mostly located in the oral and larynx (91.0%), and frequently harbor TP53 mutations (95%), as expected [53] (Figure 4A). Several lncRNAs are commonly deregulated in both cell lines and clinical samples, such as LINC01305, LINC01089, and PTOV1-AS1. There have been no detailed reports of those lncRNAs so far. The studies exploring the biological relevance and interplay between HPV infection and lncRNAs are still quite few [54,55].
TP53 mutations are the most frequent mutations in HNSCC [26,53]. We also conducted differential expression analysis between TP53 non-silent mutated tumors and TP53 wild type tumors. There were 30 down-modulated lncRNAs in TP53 mutated tumors. Among those lncRNAs, 19 lncRNAs were specifically expressed in HPV+ and TP53 wild type tumors, suggesting that these lncRNAs are potential key molecules and that their study may provide novel insights into the mechanism of HPV-associated HNSCC oncogenesis. Recently, a growing number of studies have demonstrated that lncRNAs indeed act as functional components of the p53 pathway [56]. For example, tumor suppressive lncRNA, MEG3 directly interacts with p53, and activates several p53-dependent gene transcripts [57]. p53 enhances the expression of LINC-ROR through a direct transcriptional activation. In turn, cytoplasmic-enriched LINC-ROR post-transcriptionally controls TP53 levels by sequestering hnRNP I protein and inhibiting p53 translation [58]. TUG1 has been identified as a p53 effector that is induced in response to DNA damage to mediate the inhibition of cell cycle genes [46]. Of interest, LINC00925, and LINC01315 are especially co-expressed with genes associated with p53 signaling pathways, suggesting these lncRNAs might be involved in p53 post-translational alteration and/or p53 downstream pathways. PIK3CA is also frequently mutated in HNSCC [26,53,59]. Although we performed differential gene expression analysis based on PIK3CA mutation status, no differentially expressed lncRNAs were identified in this study.
Recent studies suggest that lncRNAs are potential markers of cancer and might be helpful in the evaluation of diagnosis, classification, prognosis and distinct therapeutic strategies. In this analysis, we identified 317 lncRNAs that are expressed differentially between HNSCC tumors and normal samples, and 140 lncRNAs that differed between HPV positive tumors and HPV negative tumors. Further investigation may clarify the mechanisms underlying these differences, as well as how the changes in the HNSCC onco-lncRNAome contributes to the initiation and progression of this highly prevalent human malignancy.
Supplementary Material
Highlights.
The oral cancer onco-lncRNAome was established by bioinformatics analysis.
728 deregulated lncRNAs were identified between tumor and normal tissues.
55 lncRNAs were associated with poor prognosis.
140 transcripts were found as deregulated lncRNAs between HPV+ and HPV- specimens.
30 lncRNAs were found deregulated between TP53 mutated and TP53 wild type tumors.
Acknowledgments
This work was partially supported by the Intramural Research Program of the National Institutes of Health, National Institute of Dental and Craniofacial Research (Project: Z01DE000558), and by funds provided by the UCSD Moores Cancer Center, Office of the Dean, Medical School, UCSD (JSG), and Instituto Nacional del Cáncer, Ministerio de Salud, Argentina (MCA).
Abbreviations
- HNSCC
Head and Neck Squamous Cell Carcinoma
- HPV
Human Papilloma Virus
- lncRNA
long non-coding RNA
- TANRIC
The Atlas of Noncoding RNAs in Cancer
- TCGA
The Cancer Genome Atlas
Footnotes
CONFLICT OF INTEREST
No conflicts of interest with the contents of this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Haddad RI, Shin DM. Recent advances in head and neck cancer. N Engl J Med. 2008;359:1143–54. doi: 10.1056/NEJMra0707975. [DOI] [PubMed] [Google Scholar]
- 2.Leemans CR, Braakhuis BJM, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 3.Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol. 2007;14:103–5. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- 4.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–9. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 5.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J Pathol. 2010;220:126–39. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- 7.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–87. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quiat D, Olson EN. MicroRNAs in cardiovascular disease: from pathogenesis to prevention and treatment. J Clin Invest. 2013;123:11–8. doi: 10.1172/JCI62876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20:460–9. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 10.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A. 2008;105:716–21. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang J-R, Zhang J. Human long noncoding RNAs are substantially less folded than messenger RNAs. Mol Biol Evol. 2015;32:970–7. doi: 10.1093/molbev/msu402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12:847–65. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ebert MS, Sharp PA. Emerging roles for natural microRNA sponges. Curr Biol. 2010;20:R858–61. doi: 10.1016/j.cub.2010.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Röther S, Meister G. Small RNAs derived from longer non-coding RNAs. Biochimie. 2011;93:1905–15. doi: 10.1016/j.biochi.2011.07.032. [DOI] [PubMed] [Google Scholar]
- 18.Clark MB, Mattick JS. Long noncoding RNAs in cell biology. Semin Cell Dev Biol. 2011;22:366–76. doi: 10.1016/j.semcdb.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 19.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–61. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 20.Cheetham SW, Gruhl F, Mattick JS, Dinger ME. Long noncoding RNAs and the genetics of cancer. Br J Cancer. 2013;108:2419–25. doi: 10.1038/bjc.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li CH, Chen Y. Targeting long non-coding RNAs in cancers: progress and prospects. Int J Biochem Cell Biol. 2013;45:1895–910. doi: 10.1016/j.biocel.2013.05.030. [DOI] [PubMed] [Google Scholar]
- 22.Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339:159–66. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 23.Kunej T, Obsteter J, Pogacar Z, Horvat S, Calin GA. The decalog of long non-coding RNA involvement in cancer diagnosis and monitoring. Crit Rev Clin Lab Sci. 2014;51:344–57. doi: 10.3109/10408363.2014.944299. [DOI] [PubMed] [Google Scholar]
- 24.Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim Biophys Acta. 2014;1839:1097–109. doi: 10.1016/j.bbagrm.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 25.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.The Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–82. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Han L, Roebuck P, Diao L, Liu L, Yuan Y, et al. TANRIC: An Interactive Open Platform to Explore the Function of lncRNAs in Cancer. Cancer Res. 2015;75:3728–37. doi: 10.1158/0008-5472.CAN-15-0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin D, Abba MC, Molinolo AA, Vitale-Cross L, Wang Z, Zaida M, et al. The head and neck cancer cell oncogenome: a platform for the development of precision molecular therapies. Oncotarget. 2014;5:8906–23. doi: 10.18632/oncotarget.2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–8. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 31.Zhao Z, Bai J, Wu A, Wang Y, Zhang J, Wang Z, et al. Co-LncRNA: investigating the lncRNA combinatorial effects in GO annotations and KEGG pathways based on human RNA-Seq data. Database (Oxford) 2015;2015 doi: 10.1093/database/bav082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen G, Wang Z, Wang D, Qiu C, Liu M, Chen X, et al. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013;41:D983–6. doi: 10.1093/nar/gks1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Molinolo AA, Amornphimoltham P, Squarize CH, Castilho RM, Patel V, Gutkind JS. Dysregulated molecular networks in head and neck carcinogenesis. Oral Oncol. 45:324–34. doi: 10.1016/j.oraloncology.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quinodoz S, Guttman M. Long noncoding RNAs: an emerging link between gene regulation and nuclear organization. Trends Cell Biol. 2014;24:651–63. doi: 10.1016/j.tcb.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li D, Feng J, Wu T, Wang Y, Sun Y, Ren J, et al. Long intergenic noncoding RNA HOTAIR is overexpressed and regulates PTEN methylation in laryngeal squamous cell carcinoma. Am J Pathol. 2013;182:64–70. doi: 10.1016/j.ajpath.2012.08.042. [DOI] [PubMed] [Google Scholar]
- 37.Fang Z, Wu L, Wang L, Yang Y, Meng Y, Yang H. Increased expression of the long non-coding RNA UCA1 in tongue squamous cell carcinomas: a possible correlation with cancer metastasis. Oral Surg Oral Med Oral Pathol Oral Radiol. 2014;117:89–95. doi: 10.1016/j.oooo.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 38.Tang H, Wu Z, Zhang J, Su B. Salivary lncRNA as a potential marker for oral squamous cell carcinoma diagnosis. Mol Med Rep. 2013;7:761–6. doi: 10.3892/mmr.2012.1254. [DOI] [PubMed] [Google Scholar]
- 39.Feng J, Tian L, Sun Y, Li D, Wu T, Wang Y, et al. Expression of long non-coding ribonucleic acid metastasis-associated lung adenocarcinoma transcript-1 is correlated with progress and apoptosis of laryngeal squamous cell carcinoma. Head Neck Oncol. 2012;4:46. [Google Scholar]
- 40.Gee HE, Buffa FM, Camps C, Ramachandran A, Leek R, Taylor M, et al. The small-nucleolar RNAs commonly used for microRNA normalisation correlate with tumour pathology and prognosis. Br J Cancer. 2011;104:1168–77. doi: 10.1038/sj.bjc.6606076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zou AE, Ku J, Honda TK, Yu V, Kuo SZ, Zheng HaO, et al. Transcriptome sequencing uncovers novel long noncoding and small nucleolar RNAs dysregulated in head and neck squamous cell carcinoma. Rna. 2015;21:1–13. doi: 10.1261/rna.049262.114.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu Y, Wang J, Qiu M, Xu L, Li M, Jiang F, et al. Upregulation of the long noncoding RNA TUG1 promotes proliferation and migration of esophageal squamous cell carcinoma. Tumour Biol. 2015;36:1643–51. doi: 10.1007/s13277-014-2763-6. [DOI] [PubMed] [Google Scholar]
- 43.Han Y, Liu Y, Gui Y, Cai Z. Long intergenic non-coding RNA TUG1 is overexpressed in urothelial carcinoma of the bladder. J Surg Oncol. 2013;107:555–9. doi: 10.1002/jso.23264. [DOI] [PubMed] [Google Scholar]
- 44.Huang M-D, Chen W-M, Qi F-Z, Sun M, Xu T-P, Ma P, et al. Long non-coding RNA TUG1 is up-regulated in hepatocellular carcinoma and promotes cell growth and apoptosis by epigenetically silencing of KLF2. Mol Cancer. 2015;14:165. doi: 10.1186/s12943-015-0431-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Q, Geng P-L, Yin P, Wang X-L, Jia J-P, Yao J. Down-regulation of long non-coding RNA TUG1 inhibits osteosarcoma cell proliferation and promotes apoptosis. Asian Pac J Cancer Prev. 2013;14:2311–5. doi: 10.7314/apjcp.2013.14.4.2311. [DOI] [PubMed] [Google Scholar]
- 46.Zhang E, Yin D, Sun M, Kong R, Liu X, You L, et al. P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis. 2014;5:e1243. doi: 10.1038/cddis.2014.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cui Z, Ren S, Lu J, Wang F, Xu W, Sun Y, et al. The prostate cancer-up-regulated long noncoding RNA PlncRNA-1 modulates apoptosis and proliferation through reciprocal regulation of androgen receptor. Urol Oncol. 2013;31:1117–23. doi: 10.1016/j.urolonc.2011.11.030. [DOI] [PubMed] [Google Scholar]
- 48.Wang C-M, Wu Q-Q, Li S-Q, Chen F-J, Tuo L, Xie H-W, et al. Upregulation of the long non-coding RNA PlncRNA-1 promotes esophageal squamous carcinoma cell proliferation and correlates with advanced clinical stage. Dig Dis Sci. 2014;59:591–7. doi: 10.1007/s10620-013-2956-7. [DOI] [PubMed] [Google Scholar]
- 49.Yao J, Zhou B, Zhang J, Geng P, Liu K, Zhu Y, et al. A new tumor suppressor LncRNA ADAMTS9-AS2 is regulated by DNMT1 and inhibits migration of glioma cells. Tumor Biol. 2014;35:7935–44. doi: 10.1007/s13277-014-1949-2. [DOI] [PubMed] [Google Scholar]
- 50.Liao L-M, Sun X-Y, Liu A-W, Wu J-B, Cheng X-L, Lin J-X, et al. Low expression of long noncoding XLOC_010588 indicates a poor prognosis and promotes proliferation through upregulation of c-Myc in cervical cancer. Gynecol Oncol. 2014;133:616–23. doi: 10.1016/j.ygyno.2014.03.555. [DOI] [PubMed] [Google Scholar]
- 51.Hao Y, Wu W, Shi F, Dalmolin RJS, Yan M, Tian F, et al. Prediction of long noncoding RNA functions with co-expression network in esophageal squamous cell carcinoma. BMC Cancer. 2015;15:168. doi: 10.1186/s12885-015-1179-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hacisuleyman E, Goff LA, Trapnell C, Williams A, Henao-Mejia J, Sun L, et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol. 2014;21:198–206. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–60. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang Y, Li Y, Fang S, Jiang B, Qin C, Xie P, et al. The role of MALAT1 correlates with HPV in cervical cancer. Oncol Lett. 2014;7:2135–41. doi: 10.3892/ol.2014.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sharma S, Mandal P, Sadhukhan T, Roy Chowdhury R, Ranjan Mondal N, Chakravarty B, et al. Bridging Links between Long Noncoding RNA HOTAIR and HPV Oncoprotein E7 in Cervical Cancer Pathogenesis. Sci Rep. 2015;5:11724. doi: 10.1038/srep11724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grossi E, Sánchez Y, Huarte M. Expanding the p53 regulatory network: LncRNAs take up the challenge. Biochim Biophys Acta - Gene Regul Mech. 2015 doi: 10.1016/j.bbagrm.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 57.Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R, et al. Activation of p53 by MEG3 non-coding RNA. J Biol Chem. 2007;282:24731–42. doi: 10.1074/jbc.M702029200. [DOI] [PubMed] [Google Scholar]
- 58.Zhang A, Zhou N, Huang J, Liu Q, Fukuda K, Ma D, et al. The human long non-coding RNA-RoR is a p53 repressor in response to DNA damage. Cell Res. 2013;23:340–50. doi: 10.1038/cr.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lui VWY, Hedberg ML, Li H, Vangara BS, Pendleton K, Zeng Y, et al. Frequent mutation of the PI3K pathway in head and neck cancer defines predictive biomarkers. Cancer Discov. 2013;3:761–9. doi: 10.1158/2159-8290.CD-13-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


