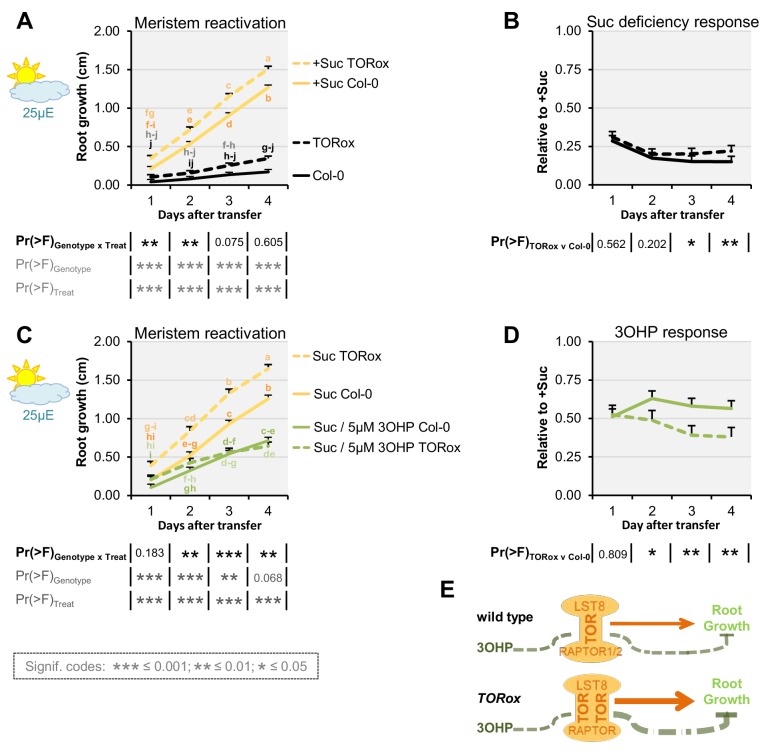
Figure 5. TOR over-activation amplifies 3OHP response.
(A) Root growth for low light grown seedlings. The seedlings were grown on MS medium without sucrose for 3 days, then transferred to the indicated media (Suc; sucrose). Multi-factorial ANOVA was used to test the impact of Genotype (Col-0 v TORox), Treatment (Control v Sucrose) and their interaction on root length. All experiments were combined in the model and experiment treated as a random effect. The ANOVA results from each day are presented in the table. (B) The root lengths grown photo-constrained and without sucrose (from A) displayed at each time point as relative to the respective sucrose activated roots. Results least squared means ± SE over three independent experimental replicates with each experiment having an average of nine replicates per condition (n = 26–30). Multi-factorial ANOVA was used to test the impact of Genotype (Col-0 v TORox), Treatment (Sucrose v Sucrose/3OHP) and their interaction on root length. All experiments were combined in the model and experiment treated as a random effect. The ANOVA results from each day are presented in the table. (C) Root growth for low light grown seedlings. The seedlings were grown on MS medium without sucrose for 3 days, then transferred to the indicated media. (D) Photo-constrained root lengths in response to sucrose and 3OHP (from A) displayed at each time point as relative to the respective sucrose activated roots. Results are least squared means ± SE over two independent experimental replicates with each experiment having an average of six replicates per condition (n = 11–14). (E) Schematic model; over expression of the catalytic subunit TOR increases growth and the relative 3OHP response.
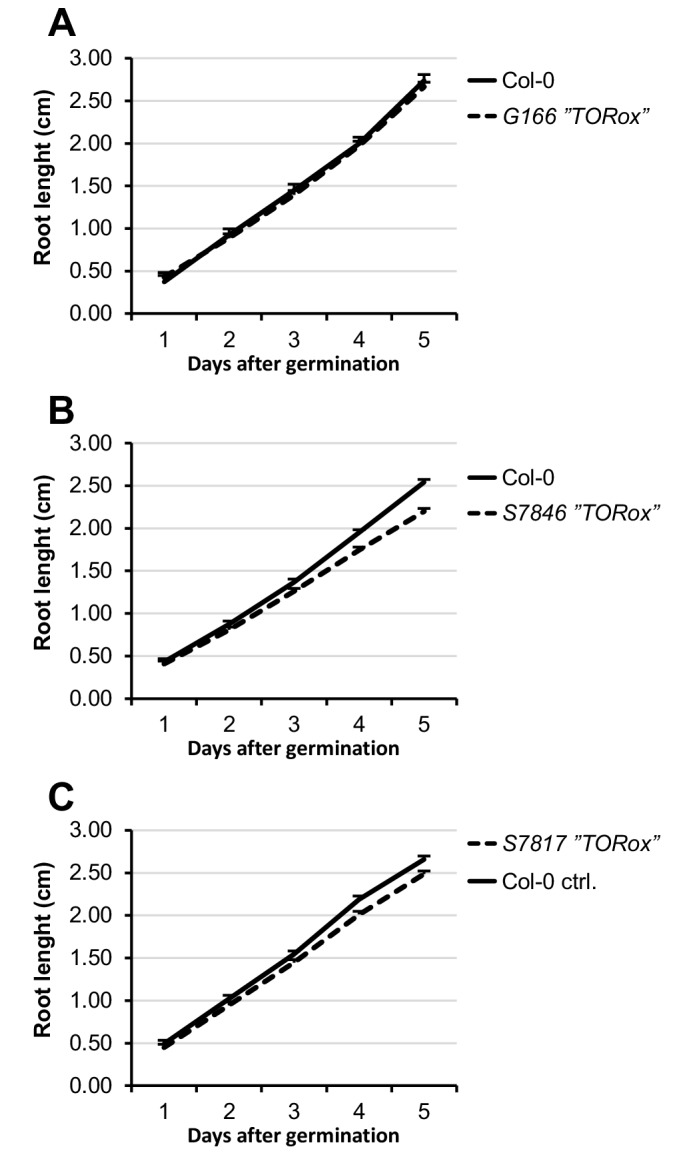
Figure 5—figure supplement 1. Published TORox lines that did not display the TORox phenotype under our conditions.