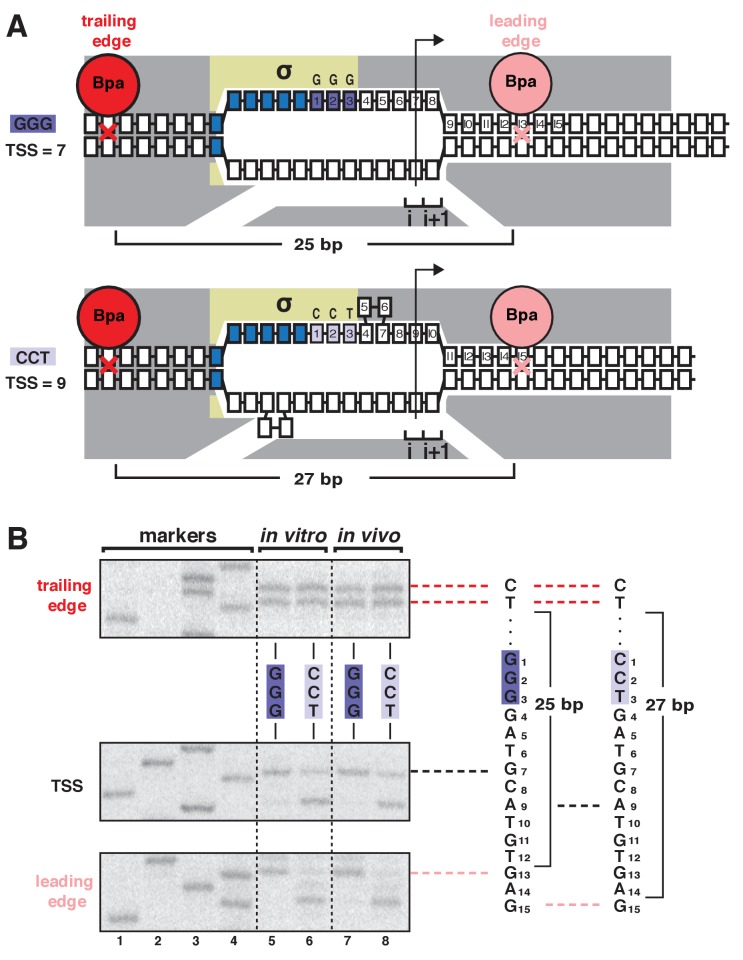
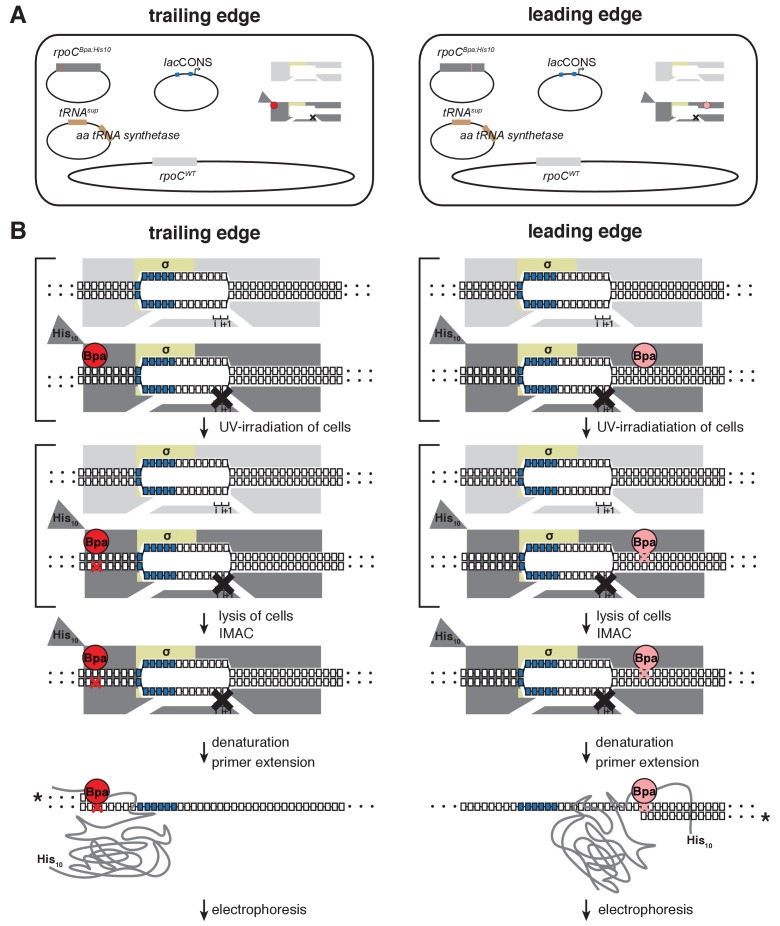
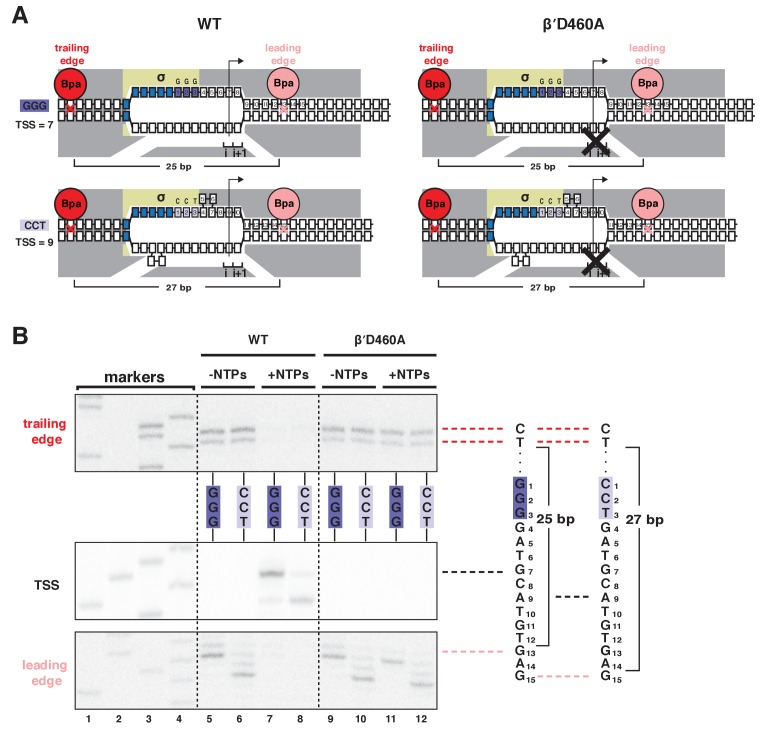
Figure 1. TSS selection exhibits first hallmark of scrunching--movement of RNAP leading edge but not RNAP trailing edge--both in vitro and in vivo.
(A) RNAP leading-edge and trailing-edge positions at promoters having GGG discriminator (TSS 7 bp downstream of −10 element; top) and CCT discriminator (TSS 9 bp downstream of −10 element; bottom). Changes in TSS selection result from changes in discriminator-sequence-dependent DNA scrunching. Gray, RNAP; yellow, σ; blue, −10-element nucleotides; dark purple, GGG-discriminator nucleotides; light purple, CCT-discriminator nucleotides; i and i + 1, NTP binding sites; arrow, TSS; boxes, DNA nucleotides (nontemplate-strand nucleotides above template-strand nucleotides; nucleotides downstream of −10 element numbered); red, trailing-edge Bpa and nucleotide crosslinked to Bpa; pink, leading-edge Bpa and nucleotide crosslinked to Bpa. Scrunching is indicated by bulged-out nucleotides. Distance between leading-edge and trailing-edge crosslinks is indicated below RNAP. (B) RNAP trailing-edge crosslinking (top), TSS (middle), and RNAP leading-edge crosslinking (bottom) for promoters having GGG discriminator and CCT discriminator, in vitro (lanes 5–6) and in vivo (lanes 7–8). Horizontal dashed lines relate bands on gel (left) to nucleotide sequences (right).