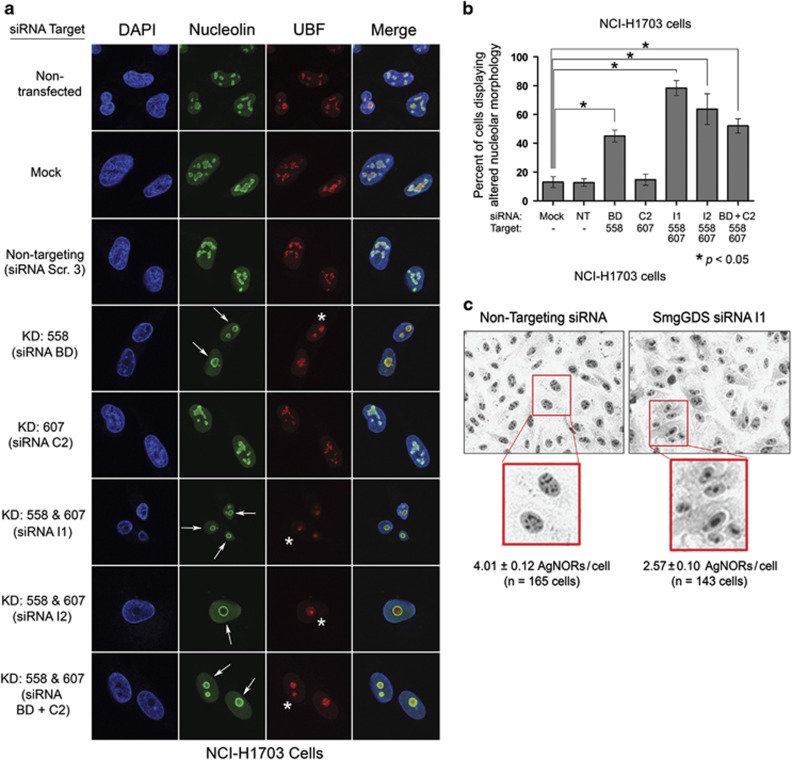
Figure 3.
The RNAi-mediated depletion of SmgGDS-558 disrupts nucleolar morphology. (a) NCI-H1703 cells were transfected with the indicated siRNAs to deplete SmgGDS isoforms, and 72 h later the cells were immunofluorescently stained with nucleolin antibody, UBF antibody and 4,6-diamidino-2-phenylindole (DAPI), and examined by confocal fluorescence microscopy. Arrows indicate circular redistribution of nucleolin, and asterisks indicate UBF cap formation (n=3). (b) NCI-H1703 cells treated the same as in a were scored for disruption of nucleolar morphology according to the key described in Supplementary Figure S3. Scoring was conducted without knowledge of the siRNAs used to treat the cells, and values represent the mean±s.e.m. from 100 cells scored in three independent experiments. Statistical significance was determined by one-way repeated measures analysis of variance and Dunnett’s multiple comparison test (*P<0.05), comparing each condition to non-targeting (NT) siRNA. (c) NCI-H1703 cells were transfected with siRNA I1 to deplete SmgGDS, or with NT siRNA, and 72 h later subjected to AgNOR staining. Phase contrast images were collected, and AgNORs/cell was counted without knowledge of the siRNAs used to treat the cells (n=3).