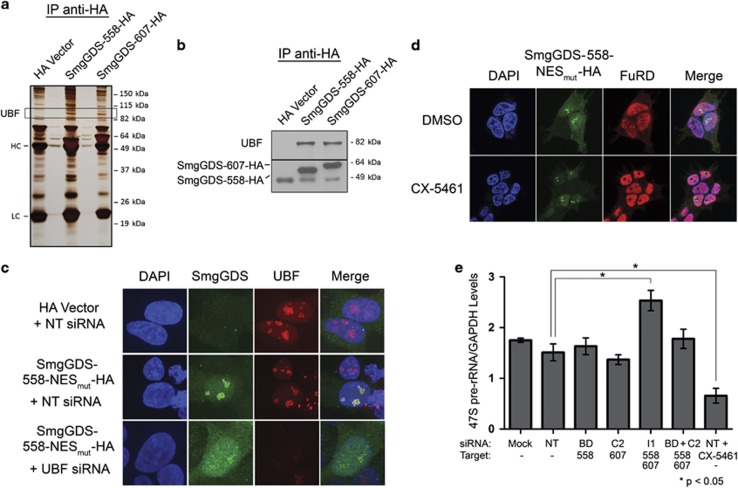
Figure 4.
SmgGDS-558 physically interacts with UBF, and this interaction promotes the nucleolar accumulation of SmgGDS-558. (a) HEK293T cells were transfected with cDNAs encoding the HA vector or HA-tagged SmgGDS, followed by immunoprecipitation using HA antibody and silver staining to detect co-precipitating proteins. Mass spectrometry identified UBF as one of the co-precipitating proteins (HC and LC; heavy and light chains, respectively, of antibodies used in the immunoprecipitation). (b) Lysates from HEK293T cells transfected with cDNAs encoding the HA vector or HA-tagged SmgGDS were immunoprecipitated using HA antibody, followed by immunoblotting using antibodies to UBF and HA (n=3). (c) HEK293T cells were transfected with cDNAs encoding the HA vector or SmgGDS-558-NESmut-HA along with non-targeting (NT) siRNA or UBF siRNA. After 72 h, the cells were immunofluorescently stained with HA antibody, UBF antibody and 4,6-diamidino-2-phenylindole (DAPI), and examined by confocal fluorescence microscopy (n=3). (d) HEK293T cells expressing SmgGDS-558-NESmut-HA were treated with or without the RNA Pol I inhibitor CX-5461 (1 μm, 2 h), followed by FuRD (2 mm, 15 min), and immunofluorescently stained using antibodies to HA and BrdU (n=3). Images were obtained by confocal microscopy. (e) NCI-H1703 cells were transfected with the indicated siRNAs to deplete SmgGDS, and 72 h later quantitative PCR was conducted to examine 47S pre-rRNA levels (normalized to cellular GAPDH). Control cells were treated with CX-5461 (1 μm, 2 h) before collecting RNA. Error bars represent ±s.e.m. of three biological replicates, and statistical significance was determined by one-way analysis of variance and Holm-Sidak multiple comparisons test (*P<0.05).