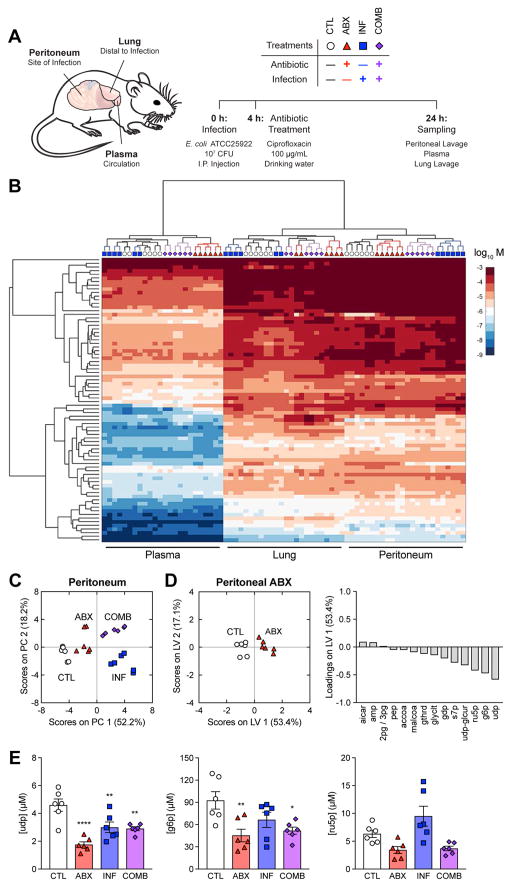
Figure 1. Antibiotic treatment depletes central metabolism intermediates in the peritoneum.
(A) Experimental design for metabolomic profiling. C57BL/6J mice were subjected to control conditions (CTL), antibiotic treatment with 100 μg/mL cipro (ABX), intraperitoneal infection with 107 CFU E. coli (INF), or their combination (COMB). Peritoneal lavage, plasma and lung lavage samples were collected 24 h after infection.
(B) Hierarchically clustered heatmap of metabolite concentrations from CTL, ABX, INF and COMB mice.
(C) PCA projection of metabolomic profiles from peritoneal samples of all four treatment groups.
(D) PLS-DA of peritoneal samples from ABX mice. Metabolites selected by elastic net regularization were depleted for central metabolism intermediates.
(E) Concentrations for metabolites with large LV1-loadings in peritoneal samples from the ABX metabolite signature. Antibiotic treatment depleted uridine diphosphate (udp), glucose-6-phosphate (g6p) and ribulose-5-phosphate (r5p).
Data are represented as mean ± SEM from n = 3 independent biological replicates. Significance reported as FDR-corrected p-values in comparison with corresponding CTL conditions: *: p ≤ 0.05, **: p ≤ 0.01, ****: p ≤ 0.0001.