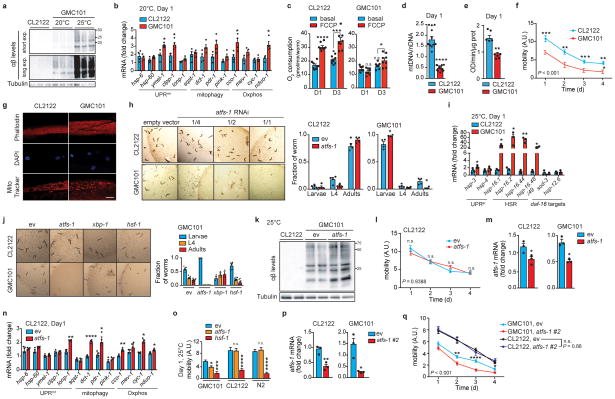
Extended Data Figure 3. Characterization of Aβ proteotoxicity and stress response pathways in GMC101 worms.
a, Amyloid aggregation in CL2122 and GMC101 worms (n=3 biologically independent samples) at 20°C or 25°C. b, MSR transcript analysis in worms at 20°C (n=3 biologically independent samples). c, Respiration assay in CL2122 and GMC101 (CL2122, n=8; GMC101, n=8 biologically independent samples). d, mtDNA/nDNA ratio in CL2122 and GMC101 (n=13 animals per group). e, CS activity in CL2122 and GMC101 on D1 (n=5 biologically independent samples). **P≤0.01 (P=0.004). f, CL2122 and GMC101 mobility (CL2122, n=48; GMC101, n=59 worms). g, Confocal images of D1 adult worms showing muscle cell integrity, nuclear morphology and mitochondrial networks. Scale bar, 10μm. See also Methods. h, Representative images and fraction of worms upon atfs-1 RNAi (n=4 independent experiments). *P<0.05 (Larvae,0.048; Adults,0.035). i, Transcript analysis of UPRer, HSR and daf-16 target genes (n=3 biologically independent samples). j, Representative images and fraction of worms fed with atfs-1, xbp-1 and hsf-1 RNAis at 20°C (n=8 per group; xbp-1, n=3 biologically independent samples). k, Amyloid aggregation upon atfs-1 RNAi (n= 2 biological replicates). l, Mobility of CL2122 fed with 50% dilution of atfs-1 RNAi (ev, n=48; atfs-11/2, n=47 worms). m, Validation of the atfs-1 RNAi in CL2122 and GMC101 (n=3 biologically independent samples). n, MSR transcript analysis of CL2122 upon atfs-1 RNAi (n=3 biologically independent samples). o, Mobility of D1 adult worms fed with atfs-1 or hsf-1 RNAi at 25°C (CL2122, n=22,27,28; GMC101, n=27,21,18; N2, n=31,38,27 worms). p, Validation of the newly generated atfs-1#2 RNAi (n=3 biologically independent samples). For further information, see Methods. q, Worm mobility upon atfs-1#2 RNAi (CL2122, ev, n=47; atfs-1#2, n=42; GMC101, ev, n=55; atfs-1#2, n=46 worms). ev, scrambled RNAi; A.U., arbitrary units. Values in the figure are mean ± s.e.m. *P<0.05; **P≤0.01; ***P≤0.001; ****P≤0.0001; n.s., non-significant. Throughout the figure, overall differences between conditions were assessed by two-way ANOVA. Differences for individual genes or two groups were assessed using two-tailed t tests (95% confidence interval). All experiments were performed independently at least twice. For uncropped gel source data, see Supplementary Fig. 1. For all the individual p values, see the Extended Data Fig. 3 Spreadsheet file.