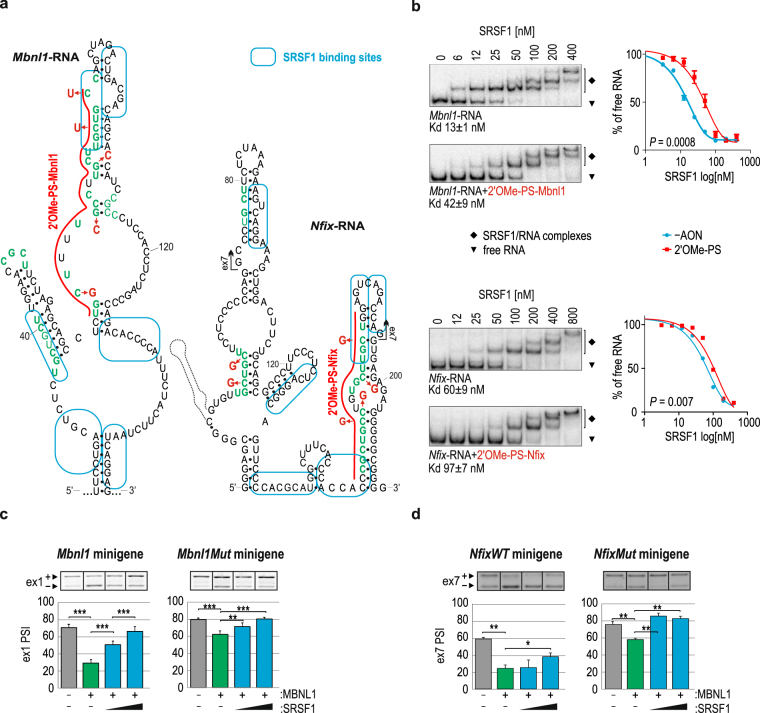
Figure 4.
Potential crosstalk between SRSF1 and MBNLs. (a) Experimentally determined secondary structures of Mbnl1-RNA and Nfix-RNA with in silico-predicted SRSF1 binding regions marked with blue boxes48,49. 2′OMe-PS AONs blocking MBNL-binding regions are marked with a red line. Point mutations are marked with red. (b) Results of EMSA showing the interaction between SRSF1 at the indicated concentrations and 5′-32P-labeled Mbnl1-RNA (upper panel) or Nfix-RNA (lower panel) with either intact MBNL-binding regions or the regions blocked by individual AONs. The calculated Kd values are indicated below each electrophoretogram; n = 4. On the right is the quantification of EMSA results based on the decline of the free RNA signal in favor of forming SRSF1/RNA complexes. Antagonistic role of SRSF1 and MBNL1 in the alternative splicing regulation of (c) ex1 of Mbnl1 and (d) ex7 of Nfix. Note the SRSF1 dose-dependent promotion of both exon inclusion into the mRNA of wild-type Mbnl1 or Nfix (left panels) or Mbnl1Mut or NfixMut minigenes (right panels). The amount of MBNL1 or SRSF1 expressing vector constituted 500 ng or ranged from 250 to 1000 ng per well, respectively; n = 2 (Mbnl1), n = 3 (Nfix).