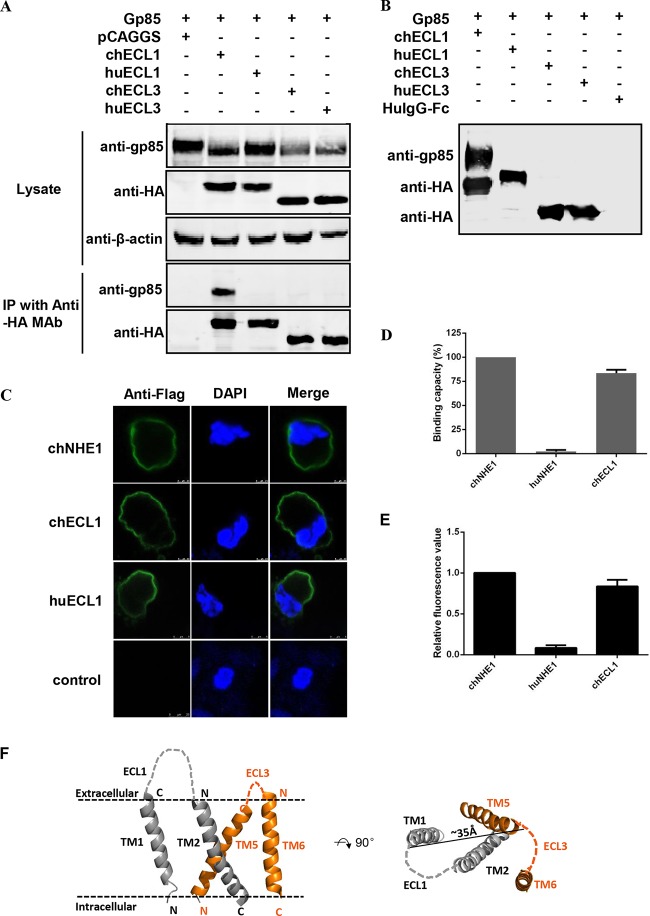
FIG 2.
chECL1 directly interacts with gp85 protein and mediates viral entry. (A) Validation of the interaction of chECL1 and chECL3 with gp85 by a coimmunoprecipitation (co-IP) assay. 293T cells were transfected with gp85- and ECL1- or ECL3-expressing plasmids. HuECL1 and huECL3 were used as a negative control. Cell lysates were prepared 48 h posttransfection, and proteins were immunoprecipitated using anti-HA-agarose MAb; the proteins were immunoblotted with 4A3 anti-gp85 MAb or anti-HA antibodies. (B) Validation of the interaction of chECL1 and chECL3 with gp85 by a pulldown assay. Soluble chECL1 and chECL3 proteins were incubated with anti-HA-agarose MAb. huECL1, huECL3, and human IgG-Fc (huIgG-Fc) were used as a negative control. The bound complexes were mixed with raw lysates of gp85 expressed by 293T cells transfected with pCAF-gp85 plasmid. The proteins were separated by SDS-PAGE and probed by immunoblotting with 4A3 anti-gp85 MAb or anti-HA antibodies. (C) Confocal microscopy analysis of the cell surface expression of ECL1 constructs in 293T cells. 293T cells were transfected with chECL1 and huECL1 expression plasmids for 24 h and fixed and stained with anti-FLAG M2 MAb, followed by incubation with Alexa Fluor 488 donkey anti-mouse IgG(H+L) (green). Nuclei were stained with DAPI. chNHE1 was used as a positive control. (D) The gp85-binding capacity of chECL1 and huECL1 expressed on the surface of transfected 293T cells, evaluated in a receptor binding assay. (E) Entry of RCAS(J)-luciferase virus into 293T cells expressing chECL1 or huECL1, assayed as described in the legend to Fig. 1. (F) Modeling of the chNHE1 transmembrane (TM) domain. For clarity, only TM domains of ECL1 (TM1 and TM2) and ECL3 (TM5 and TM6) are shown. Cell membranes are indicated by dotted lines. The distance between C-terminal portions of TM1 and TM5 is shown to indicate the size of the molecule. The structural modeling of the receptors was generated using SWISS-MODEL and was based on homology molecules found in the PDB database. (D and E) Three independent experiments were performed, and data are shown as means ± standard deviations for triplicates from a representative experiment.