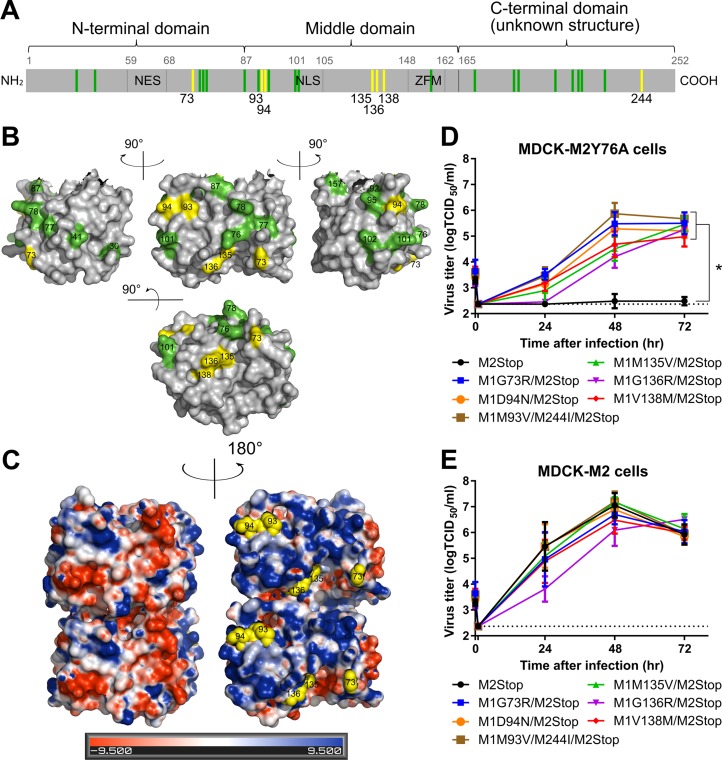
FIG 2.
M1 suppressor mutations complement the M2Y76A defect in virus replication. The M gene of viruses with suppressor mutations for M2Y76A was sequenced, and rUd-M2Stop viruses with M1 suppressor mutations were generated by reverse genetics. (A) The identified suppressor mutations in M1 protein (yellow) and residues known to affect virus morphology/assembly (green) are shown (13–19). The nuclear export sequence (NES), nuclear localization sequence (NLS), and the zinc finger motif (ZFM) are also indicated. (B) M1 suppressor mutations (yellow) and known residues that affect virus morphology (green) are shown on the M1 crystal structure (aa 1 to 164, Protein Data Bank [PDB] accession no. 1EA3 [21]) using PyMOL. (C) M1 suppressor mutations (yellow spheres) are located on the positively charged surface (blue) and not the negatively charged surface (red) of the M1 dimer neutral-pH structure (PDB accession no. 1EA3). Electrostatic surface potentials of M1 were generated by the PDB2PQR server and PyMOL APBS tools. (D and E) rUd-M2Stop virus and rUd-M2Stop viruses with M1 suppressor mutations were used to infect MDCK-M2Y76A cells (D) or MDCK-M2 cells (E) at an MOI of 0.001. Infectious virus titers in the cell supernatants were quantified using TCID50 assays. The dotted lines indicate the limits of detection. Asterisks indicate statistically significant differences compared to rUd-M2Stop-infected cells (P < 0.01 using two-way multiple-comparison ANOVA with Bonferroni posttest). Data were analyzed from three independent experiments. Error bars are standard deviations.