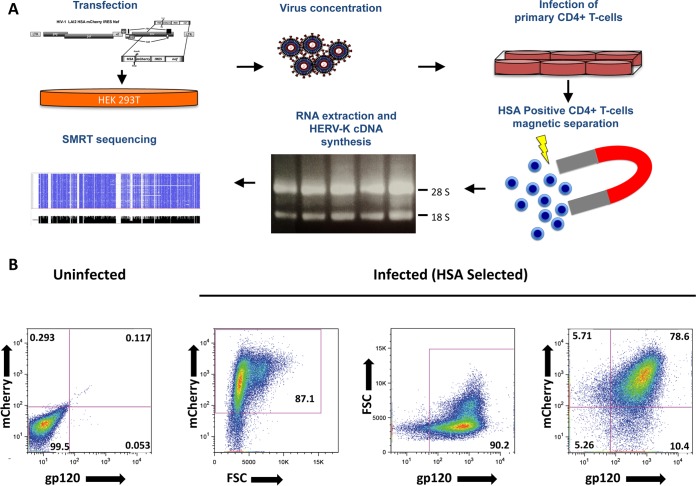
FIG 2.
(A) Stepwise schematic representation of the procedure leading to the enrichment of infected CD4+ T cells necessary to achieve a low-background HIV-1-defined HERV-K expression profile. Virus production includes transfection of HEK293T and viral supernatant concentration/purification over density barrier layer (OptiPrep/odixanol). Activated CD4+ T cells are then infected overnight and incubated after a washing step for an additional 3 days. Separation of HSA-positive cells includes incubation with biotin-conjugated α-mCD24 (HSA), followed by the addition of streptavidin-coated magnetic nanoparticles, and magnetic separation. After washing, the cells are lysed in TRIzol, and the total RNA is extracted. The RNA is then used to generate HERV-K HML-2 expression cDNA libraries that are analyzed by long-read SMRT sequencing. (B) More than 90% of the HSA enriched are HIV-1 infected. At 4 days after infection with LAI2 HSA-mCherry-IRES-Nef, the cells magnetically separated for HSA were stained with α-gp120 human antibody (clone 2G12) and analyzed by flow cytometry. Dot plots show the frequencies of cells positive for mCherry expression and anti-gp120 staining. FSC, forward scatter.