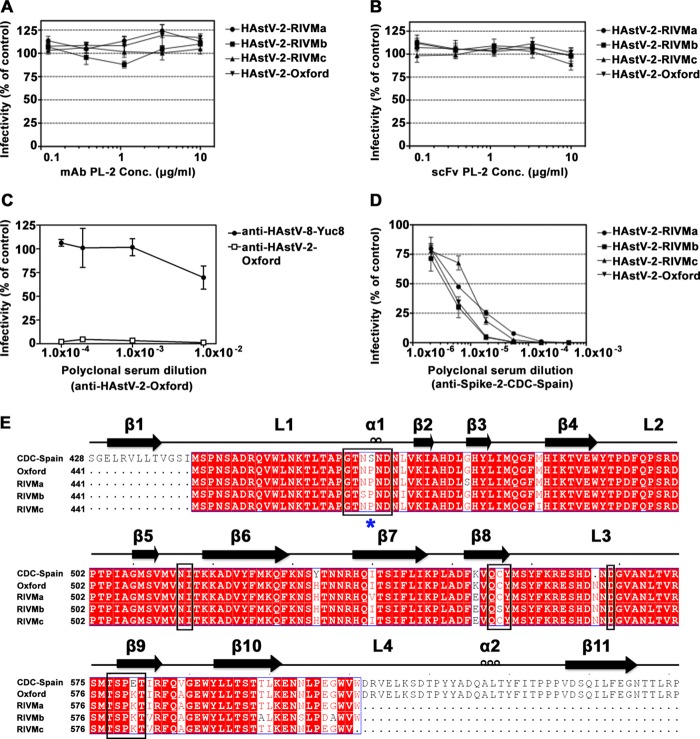
FIG 1.
Antibody neutralization of HAstV-2 strains and alignment of HAstV-2 capsid spike sequences. (A and B) Infectivity of indicated HAstV-2 strains preincubated with MAb PL-2 (A) and with scFv PL-2 (B). (C) Infectivity of HAstV-2-Oxford preincubated with anti-HAstV-2-Oxford or anti-HAstV-8-Yuc8 polyclonal sera. (D) Infectivity of indicated HAstV-2 strains preincubated with anti-Spike-2-CDC-Spain polyclonal sera. All infectivity experiments were performed in biological triplicates, and the error bars represent the standard errors of the means. (E) Sequence alignment of Spike-2-CDC-Spain, Spike-2-Oxford, and Spike-2-RIVMa-c. Conserved, similar, and nonconserved amino acids are colored red, pink, and white, respectively. Alignments and mapping of conservation onto the structure were performed with the online ENDscript server. Black boxes highlight amino acids in the antibody PL-2 epitope. A blue star indicates the location of Ser463 or Pro463.