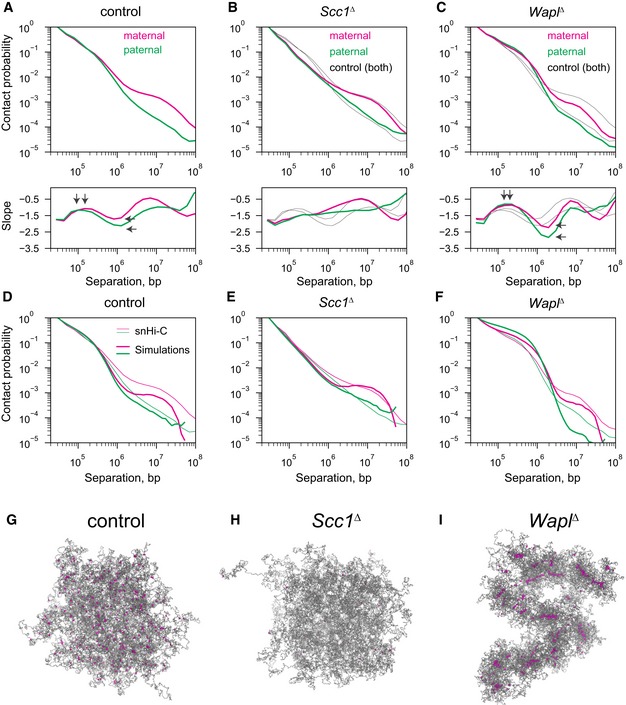
Figure 4. Differences in genome‐wide contact probability, Pc(s), for chromatin loci separated by genomic distances, s, between conditions.
-
A–CExperimental Pc(s) for maternal and paternal chromatin for Scc1 control, Scc1 ∆, and Wapl ∆ conditions. Black solid lines in (B and C) show the control curves as a reference to guide the eye. Slopes of the log(Pc(s)) curves for each condition are shown in the subpanel below each Pc(s) plot. Vertical arrows on the slope subpanels indicate the maximum slope, which is used to infer the average size of cohesin‐extruded loops; this analysis indicates that the average extruded loop size is approximately 60–70 kb in control zygotes and increases in the Wapl ∆ condition to over 120 kb. Horizontal arrows on the slope panels indicate the minimum slope, which can indicate cohesin linear density on the chromatin; notably, neither maternal nor paternal Scc1 ∆ zygotes have a minimum slope suggesting very low cohesin density, whereas minima exist in both control and Wapl ∆ conditions. Data are based on n(Wapl fl, maternal) = 7, n(Wapl fl, paternal) = 6, n(Wapl ∆ , maternal) = 8, n(Wapl ∆ , paternal) = 7, n(Scc1fl, maternal) = 13, n(Scc1 fl, paternal) = 17, n(Scc1 ∆, maternal) = 28, and n(Scc1 ∆, paternal) = 17 nuclei, from at least two independent experiments using two to three females per genotype each.
-
D–FSimulated chromatin Pc(s) for the control, Scc1 ∆, and Wapl ∆ conditions. Simulation Pc(s) curves shown in thick lines and experimental Pc(s) curves in thin lines.
-
G–IRepresentative images of the simulated paternal chromatin fiber used for the Pc(s) calculations in panels (D–F). The chromatin fiber is colored in gray, and the locations of the cohesins are colored in purple.
Source data are available online for this figure.
