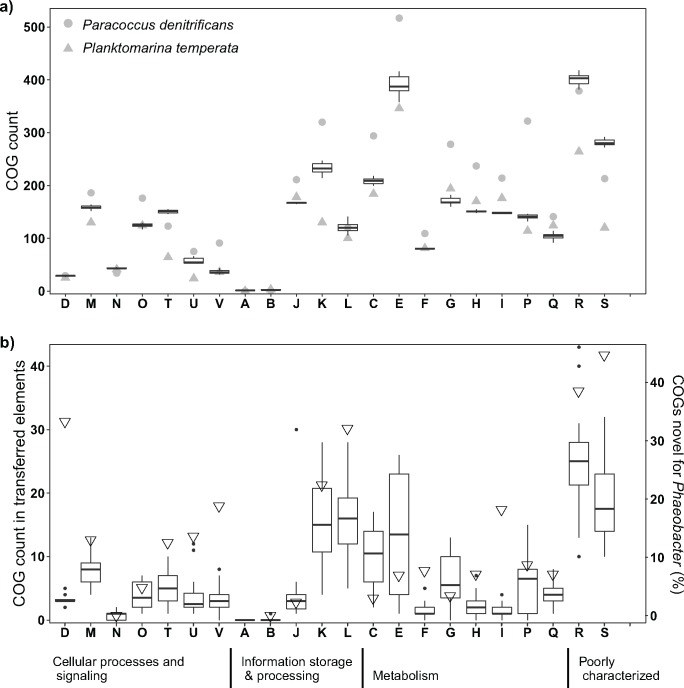
Fig. 3.
—Abundance of different COGs present in the 32 Phaeobacter chromosomes in comparison to Paracoccus denitrificans PD1222 and Planktomarina temperata RCA23, DSM 22400, both from IMG, sorted by categories (a) and number of COGs gained through HGT (b). Open triangles indicate percentage of COGs gained by HGT that are novel for the genus Phaeobacter. Box plots show median, 25% and 75% percentiles, whiskers the 1.5*interquartile range and all values outside the range are shown as outliers. Letters indicate COG categories: A (RNA processing and modification), B (chromatin structure and dynamics), C (energy production and conversion), D (cell cycle control, cell division, chromosome partitioning), E (amino acid transport and metabolism), F (nucleotide transport and metabolism), G (carbohydrate transport and metabolism), H (coenzyme transport and metabolism), I (lipid transport and metabolism), J (translation, ribosomal structure and biogenesis), K (transcription), L (replication, recombination and repair), M (cell wall/membrane/envelope biogenesis), N (cell motility), O (posttranslational modification, protein turnover, chaperones), P (inorganic ion transport and metabolism), Q (secondary metabolites biosynthesis, transport and catabolism), R (general function prediction only), S (function unknown), T (signal transduction mechanisms), U (intracellular trafficking, secretion, and vesicular transport), and V (defense mechanisms).