Fig. 1.
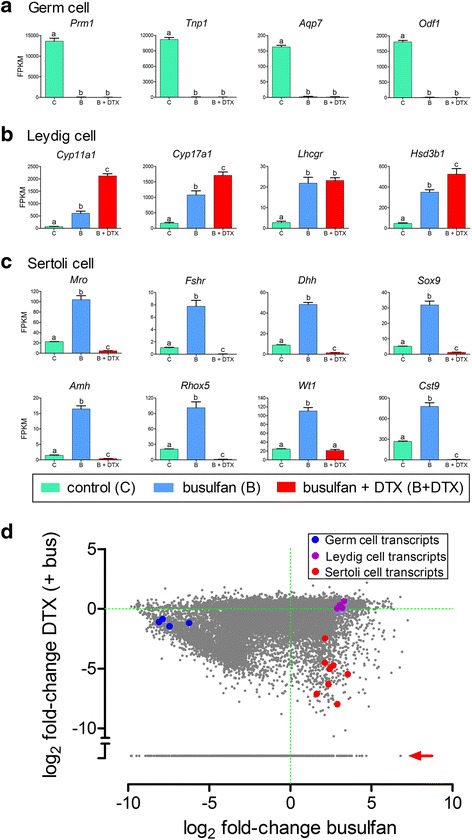
Data from RNAseq study showing transcript expression in control, busufan-treated and busulfan + DTX-treated mice. Data shows expression of known germ-cell transcripts (a), Leydig cell transcripts (b) and Sertoli cell transcripts (c). Results show mean ± SEM of 4 or 5 animals per group. In (d) data from all transcripts detected in the RNAseq study is shown as the transcript number log ratio of DTX + busulfan/ busulfan alone (log2 fold-change DTX (+bus)) plotted against the log ratio of busulfan alone/control (log2 fold-change busufan). Known germ cell transcripts from a) are shown in blue, Leydig cell transcripts from b) are shown in purple and Sertoli cell transcripts from c) are shown in red. When transcript expression in the DTX + busulfan group was zero the log2 fold-change DTX (+bus) could not be calculated and the points are indicated by a red arrow. In a) to c) groups with different letter superscripts are significantly (P < 0.05) different
