Abstract
Fibroblast growth factors (FGF) are essential signaling proteins that regulate diverse cellular functions in developmental and metabolic processes. In Drosophila, the FGF homolog, branchless (bnl) is expressed in a dynamic and spatiotemporally restricted pattern to induce branching morphogenesis of the trachea, which expresses the Bnl-receptor, breathless (btl). Here we have developed a new strategy to determine bnl-expressing cells and study their interactions with the btl-expressing cells in the range of tissue patterning during Drosophila development. To enable targeted gene expression specifically in the bnl expressing cells, a new LexA based bnl enhancer trap line was generated using CRISPR/Cas9 based genome editing. Analyses of the spatiotemporal expression of the reporter in various embryonic stages, larval or adult tissues and in metabolic hypoxia, confirmed its target specificity and versatility. With this tool, new bnl expressing cells, their unique organization and functional interactions with the btl-expressing cells were uncovered in a larval tracheoblast niche in the leg imaginal discs, in larval photoreceptors of the developing retina, and in the embryonic central nervous system. The targeted expression system also facilitated live imaging of simultaneously labeled Bnl sources and tracheal cells, which revealed a unique morphogenetic movement of the embryonic bnl- source. Migration of bnl- expressing cells may create a dynamic spatiotemporal pattern of the signal source necessary for the directional growth of the tracheal branch. The genetic tool and the comprehensive profile of expression, organization, and activity of various types of bnl-expressing cells described in this study provided us with an important foundation for future research investigating the mechanisms underlying Bnl signaling in tissue morphogenesis.
Keywords: branchless, breathless, CRISPR/Cas, LexA/lexO, neurons, tracheoblast
1. Introduction
FGF family of signaling proteins and their receptors are essential to regulate broad range of functions in metazoan development and in adult tissue homeostasis. Abnormal FGF signaling is linked to many congenital disorders as well as cancers (Beenken and Mohammadi, 2009; Brewer et al., 2016; Turner and Grose, 2010). Understanding the fundamental basis of FGF signaling is the key to uncover mechanisms of development and diseases. The Drosophila tracheal branching morphogenesis is an excellent model to investigate the dynamics of intercellular FGF communication underlying tissue patterning. The tracheal branching morphogenesis is regulated by paracrine signaling activity of bnl, one of the three fgf genes in Drosophila (Bae et al., 2014; Stathopoulos et al., 2004; Sutherland et al., 1996). Dynamic and spatiotemporally regulated expression of Bnl signal is known to guide the directed migration of tracheal branches toward the signal source. For instance, in the embryo, bnl expression in five clusters of epidermal cells surrounding the tracheal placode controls budding and growth of five different primary branches. In 3rd instar larva, bnl expression in a small group of wing imaginal disc cells induces growth of a tracheal branch, the air sac primordium (ASP), a precursor of the adult dorsal air sac (Sato and Kornberg, 2002). Similarly, during pupal metamorphosis, decaying larval tracheal branches express bnl, which guides the migration of tracheal progenitors along a preexisting scaffold of dying tracheal cells to replace old tissue (Chen and Krasnow, 2014). Genetic evidences suggested that Bnl-signaling is also important for several non-tracheal morphogenesis events. These functions include control of cell adhesion in the embryonic midline epithelia and in the developing retina, cell proliferation in larval neuroblasts, migration of glial cell in the embryonic central nervous system (CNS) and germ cells, and the mesenchymal-epidermal transition in male larval genital discs (Ahmad and Baker, 2002; Barrett et al., 2008; Klämbt et al., 1992; Mukherjee et al., 2012).
Despite the important roles of Bnl, it is unknown how its expression and activity are dynamically controlled during tracheal morphogenesis. The identity, organization and functional interactions of local populations of bnl-source that influence in germ cell migration, midline epithelia, embryonic CNS, and in larval eye disc have not been characterized. Therefore, to understand the fundamental mechanisms of bnl signaling, at first a comprehensive idea of the organization and interaction of bnl- and btl-expressing cells in various cellular and developmental contexts is necessary. Currently, the most reliable assessment of bnl expression is obtained from the RNA in situ hybridization and immunostaining using a bnl-lacZ enhancer trap line. However, the subcellular localization of signals from bnl in situ hybridization and bnl-lacZ immunostaining limits the investigation of cellular morphology during development. These fixed tissue methods also limit documentation of dynamic changes in the live embryos. Moreover, to investigate the mechanism of signaling interaction between btl-and bnl-expressing cells, available genetic tools (Fig.1) are incapable of simultaneously labeling and manipulating gene activities in the Bnl exchanging cells.
Figure 1. Expression profile of existing bnl enhancer trap lines. A-C″.
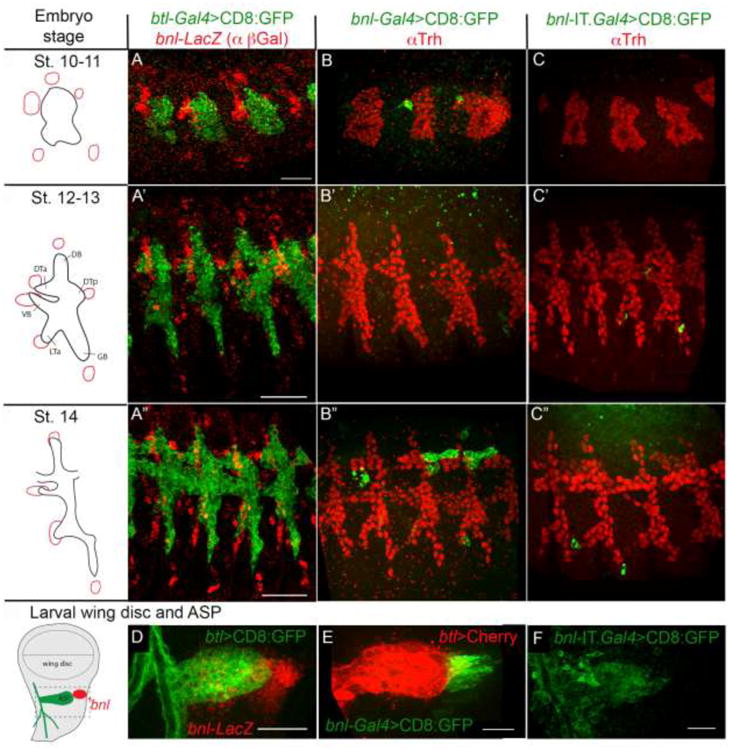
Embryonic expression of three bnl enhancer trap lines: bnl-lacZ (red, pGal antibody; A-A′), bnl-Gal4 (B-B′), and bnl-IT.Gal4 (C-C). D-F, Expression of bnl-lacZ (red, D), bnl-Gal4 (E) and bnl-IT.Gal4 (F) in the larval wing imaginal discs and relative location of the bnl expression with the Air Sac Primordium (ASP). Tracheal cells were marked by either CD8:GFP (A-A″ and D; btl-Gal4 X UAS-CD8:GFP) or Trh antibody staining (B-C″; red), or btl-LexA (E; btl-LexA X lexO-CherryCAAX). F, non-specific expression of bnl-IT.Gal4 in larval ASP (bnl-IT.Gal4 X UAS-CD8:GFP). Left column, schematic drawings of the bnl expression patterns (red) at the embryonic developmental stages and in the larval wing discs obtained in earlier studies (Sato and Kornberg, 2002; Sutherland et al., 1996). Scale bars, 30μm.
To achieve these goals, here we generated a targeted expression system specific for bnl expressing cells using the Clustered Regularly Interspaced Short Palindromic Repeat/CRISPR associated (CRISPR/Cas)- mediated genome editing (Jinek et al., 2012; Bibikova et al., 2002). This genome editing technique utilized two small chimeric guide RNAs (gRNA), which directed the bacterial endonuclease Cas9 to create double-strand breaks (DSB) at defined positions to delete the first coding exon of bnl. The DSBs were subsequently repaired by homologous recombination (HR) to swap the bnl exon with the sequence of the bacterial transactivator LexA. In the new expression system generated here, endogenous bnl regulatory sequences in the genome control the expression of LexA, which, in turn, binds to the LexA operator element, lexO, and activates expression of any transgene placed under lexO control (Lai and Lee, 2006). Analyses of bnl-LexA, had identified previously uncharacterized expression, organization and functional interaction between the bnl- and btl- expressing cells in tracheal and neuronal morphogenesis. The novel tool and the new expression profiles described in this study are important resources that should rapidly advance future investigations of FGF signaling and their interpretation.
2. Materials and Methods
2.1. CRISPR/Cas9 based genome editing
Guide RNA design and construction
Two gRNA target sites were selected within the first coding exon of the bnl using the flyCRISPR Optimal Target Finder tool and were designed following the recommendations ((Gratz et al., 2014), Figure 2A). All the candidates were evaluated by setting “maximum stringency” and “NGG only” for “PAM”. Two sites with no potential off-targets were selected and sequence verified to ensure no mutations in the genome of the parent fly stocks selected for injection (nos-Cas9 (on X chromosome), BL# 54591). Also, the potentially “good” activities of both gRNAs were predicted using the software tool described in (Doench et al., 2014). Two gRNAs that matched all those criteria (PAM sequence underlined) were:
Figure 2. Generation of bnl-LexA using CRISPR/Cas9 based genome editing. A.
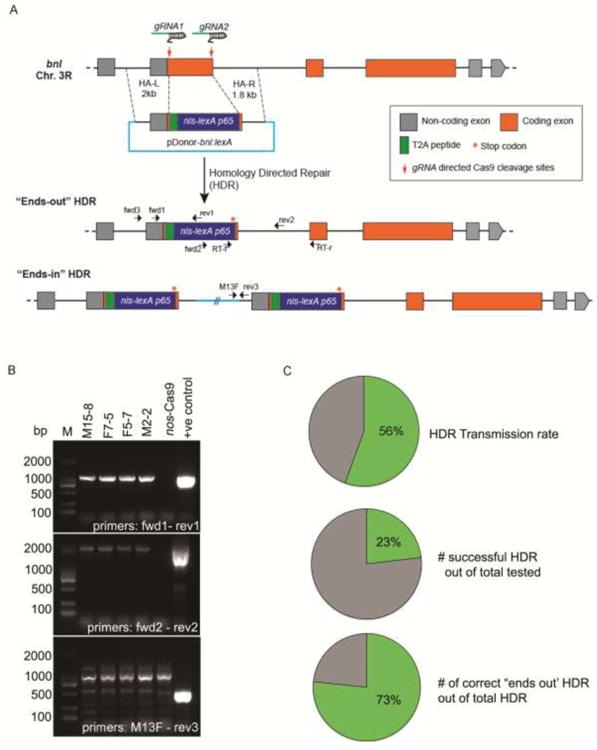
Schematic representation of the CRISPR/Cas9 mediated HDR strategy for exon replacement in the bnl locus. Box - exon, orange - coding region of the exon; grey - non-coding region of the exon; line- intron; red arrows - gRNA target sites; donor plasm id (pDonor-bnllexA) and two possible outcomes of the engineered loci were shown. The pDonor-bnllexA contained: (i)T2A-nls-lexA:p65 (∼1.8kb) sequence flanked by about 2 kb homology arms containing the genomic sequences from the 5′ and 3′ regions to the cleavage sites (dashed lines), (ii) T2A self-cleaving peptide between a residual N terminal bnl exon and the nls-lexA:p65, and a translation stop codon (red *) immediately after the nls-lexA:p65 sequence. The HDR product is expected to disrupt bnl function, make a chimeric bnl-LexA mRNA under the same transcriptional and post-transcriptional control, and yield only the LexA:p65 protein. Small black arrows represent relative locations (not in scale) of the PCR primers (Table 2, primers # 9-16) used for screening or RT-PCR. B, photograph of agarose gel electrophoresis showing results of the three-step PCR based screening strategy. PCR products amplified from the genomic DNA of four successful ends-out HDR lines shown. Negative control, PCR product amplified from the genomic DNA of nos-Cas9 parental line; positive control, pDonor-bnllexA plasmid; M, Marker (SL2K DNA ladder). C, efficiency of the genome editing shown in pie graph (also see Table 1).
gRNA1: TGTATCTGCGATGCCCCTCATGG gRNA2:
ATCCTTCAGATATTGCGGGATGG
These gRNAs were expected to delete a 835 nucleotide (nt) region of the coding exon. The gRNAs were cloned using Gibson Assembly (NEB) of the PCR products (amplified with primers listed in Table 2) into a pCFD4 RNA expression vector following a gRNA cloning protocol (Port et al., 2014). All PCR reactions were carried out either with KAPA HiFi Hot Start- (Kapa Biosystems) or Q5 Hot Start High Fidelity- (NEB) DNA polymerase following the manufacturers' protocol. For gel purification or clean-up of the PCR products, purification kits from Zymo Research were used.
Table 2. Primers used in this study.
| # | Name | Sequence |
|---|---|---|
| 1 | bnl-lexA gRNA fwd | TATATAGGAAAGATATCCGGGTGAACTTCgTGTATCTGCGATGCCCCTCAGTTTTAGAGCTAGAAATAGCAAG |
| 2 | bnl-lexA gRNA rev | ATTTTAACTTGCTATTTCTAGCTCTAAAACTCCCGCAATATCTGAAGGATcGACGTTAAATTGAAAATAGGTC |
| 3 | bnl N-F_pUC19 | AATTCGAGCTCGGTACtgtggtctttgaggctggaac |
| 4 | bnl-lexA-N-R | tCCGcaagtCagtAGgctgccgcgtccttcgccggaGCCCGCAGATACAAGGCCCC |
| 5 | lexA-F | CTactGacttgCGGaGAtGTcGAaGAGAACCCtGGCCCtATGCCACCCAAGAAGAAGC |
| 6 | lexA-R | CTAAACGAGTTTTTAAGCAAACTCACTC |
| 7 | bnl lexA-C Fwd | TAAAAACTCGTTTAGACGGGATGGCGTTGTCAAC |
| 8 | bnl C-R_pUC19 | GCCAAGCTTGCATGCCtcgcataattgccgcctgg |
| 9 | bnl-lexA scr fwd1 | GTGGCGCACGCCCAATAAAC |
| 10 | bnl-lexA scr rev1 | GATCCCAGCCAATCTCCGTTG |
| 11 | bnl-lexA scr fwd2 | CAACGGAGATTGGCTGGGATC |
| 12 | bnl-lexA scr rev2 | CTGGCCAACTGTAGGGAAGTC |
| 13 | ends-in check rev3 | GCAATGTTATGCAATGCGTTGAC |
| 14 | bnl-lexA seq fwd3 | CACTTGTCGCCCATATTGATACAATTG |
| 15 | lexA primer 5F | GATATGGATTTCTCCGCTTTGCTG |
| 16 | FGF domain R2 | CCATGCAGAGATACAGGCAAGTG |
Primer #1,2: were used for gRNA cloning; nucleotides underlined anneal to U6 promoter or gRNA core on pCFD4 vector, the lowercase g or c was added to aid transcription by the U6 promoter. Primer # 3, 4, 5, 6, 7, 8: for replacement donor construction, nucleotides in capital from # 3 or #8 overlap with pUC19 vector for Gibson Assembly, nucleotides underlined in # 4 or #5 were sequence overhang for T2A peptide addition. Primer # 9-14: were used for CRISPR screening and sequencing and were shown in Fig 2 as fwd1-3 and rev1-3. Primer # 15, 16: were used for RT-PCR analyses and were shown in Figure 2 as RT-f and RT-r.
Replacement donor design and construction
The replacement donor was designed to contain the T2A-nls-lexA:p65 sequence flanked by 5′ (2 kb) and 3′ (1.8 kb) homology arms. Long homology arms were selected for efficient homology directed repair (HDR) and insertion of a large 1.8 kb T2A-nls-lexA:p65 fragment at the site of repair. Longer homology arms are known to increase the chance of HDR during the repair process (Beumer et al., 2013). The 5′ homology arm contained the UTR and the 93nt coding region of the engineered coding exon. The 3′ arm contained the 46nt of the 3′ part of the same coding exon along with the downstream bnl intronic sequence. For efficient translation of nls-lexA:p65 as a separate protein product from the chimeric bnl-lexA mRNA, a T2A sequence was incorporated between the 93 nt 5′ bnl coding region and the nls-lexA:p65. A translation stop codon was incorporated after the nls-lexA:p65 sequence. Both homology arms were amplified from genomic DNA extracted from the targeted fly line (nos-Cas9) using primers in Table 2. The nls-lexA:p65 sequence was amplified from pBPnlsLexA:p65Uw vector (Addgene) using primers in Table 2. Gibson assembly was used to clone the three fragments together in the pUC vector to generate the replacement construct, pDonor-bnl:lexA. The cloned region was fully sequenced before being used for genomic replacement. For plasmid purification, GeneJET Miniprep kit and PureLink® HiPure Plasmid Maxipep kits (ThermoFisher Scientific) were used. Both gRNA recognition sites were disrupted in the replacement donor by the exogenous lexA cassette. The disrupted gRNA recognition sites in the replacement cassette (the exogenous sequences that disrupted gRNA binding sites shown in italics) were:
gRNA1: TGTATCTGCG-GGCTCCGGCGAAGGACnnn
gRNA2: nnnAAAAACTCGTTTAGA-CGGGATGG
About 835 bp middle region of the first bnl-coding exon was replaced by the nls-lexA:p65 coding sequence.
Embryo injection, fly genetics and PCR based screening
The parent, nos-Cas9 (on X chromosome) embryos were injected with 100ng/μL of pCFD4-gRNA and 500ng/μL of pDonor-bnllexA (Rainbow Transgenic Flies Inc). When the injected embryos developed into adults, single G0 founders were crossed to 3rd chromosome balancer flies. 20 male offspring (F1 generation) from each fertile cross were individually crossed to the balancer females. When F2 larvae hatched, the single F1 father from each cross was sacrificed for genomic DNA extraction. DNA was also extracted from a nos-Cas9 fly, which served as a negative control. PCRs were performed using the primers indicated in Figure 2A, Table 2. PCRs using primers fwd1 and rev1 were performed to screen for the LexA replacement, and PCRs using fwd2 and rev2 primers were performed to check the 3′ region. PCRs using primers M13F and rev3 were performed to check “ends-in” HDR. The balanced stocks (over TM6) were established from the F2 flies. High quality genomic DNA was prepared from the established stocks for full sequencing of the engineered regions. Out of total 42 lines that showed expression consistent with the previously characterized bnl patterns, 10 lines were randomly selected and confirmed by sequencing. Two completely sequenced lines were used in all analyses.
2.2. Drosophila strains
The following fly strains were obtained from Bloomington Stock Center: bnl-lacZ (bnlP2), bnl- IT.Gal4, elav-GAL4, eg-GAL4, UAS-CD8:GFP, UAS-CD8:RFP, lexO-CD8:Cherry, and UAS-CD4:IFP2, lexO-nsyb:GFP1-10, UAS-CD4:GFP11, His2Av-mRFP1, UAS-λBtl, UAS-Btl-DN. bnl-LexA was created in this study. bnl-Gal4 and btl-Gal4 were described earlier (Roy et al., 2014). For nsyb-GRASP experiment, lexO-nsyb: GFP1-10 UAS-CD4:GFP11; UAS-CD8:RFP virgin females were crossed to btl-Gal4; bnl-LexA/TM6 males. All crosses were incubated at 25°C.
2.3. Microscopy
Live and fixed cell imaging were performed using either a spinning disc confocal microscope (Andor and Perkin Elmer) or a scanning confocal microscope (Leica SP5X). For time lapse imaging of the developing trachea and bnl source, dechorionated embryos were imaged through glass bottom petri dishes (MatTek), over 60 μm z-stacks, every 10 minutes for a duration of ∼5-6 hours in an Andor spinning disc microscope. Cells in embryo were tracked using the Tracking plugin in Fiji. Subsequent analyses of migration of the dorsal- and lateral- bnl-expressing clusters were carried out with Chemotaxis and Migration Tool 2.0 from ibidi. In situ hybridization images were acquired with an upright Nikon Eclipse Ni microscope and a color CCD camera. The images were analyzed with iQ3 and Fiji software.
2.4. Immunostaining
Drosophila embryos were collected, fixed and rehydrated by standard protocols. Larval tissues were fixed for 25 min at room temperature in 4% paraformaldehyde (PFA) in PBS and washed in PBS with 0.1% Triton X-100 (PBST). When immunostaining was not required, following a wash in PBS, specimens were directly mounted on slides in VECTASHIELD medium. For immunostaining, antigen blocking was carried out at room temperature for 30-60 min in 5% Normal Goat Serum (NGS)/PBST. Primary antibody incubations were carried at 4°C overnight with gentle rocking. The following antibodies were used in this study: mouse anti-Prospero (1:10, DSHB), rat anti-Trh (1:200, a gift from Dr. Lan Jiang), rat anti-Elav (1:100, DSHB), mouse anti-Repo (1:100, DSHB), rat anti-DCAD2 (1:10, DSHB), guinea pig anti-Dpn (1:1000, a gift from Dr. James Skeath), guinea pig anti-Hb9 (1:1000, a gift from Dr. James Skeath), guinea pig anti-Ems (1:300, a gift from Dr. Haluk Lacin), mouse anti-Engrailed (1:50, DSHB), mouse anti-Cut (1:50, DSHB), guinea pig anti-Senseless (1:1000, a gift from Dr. Hugo Bellen), mouse anti-Hemese (1:100, a gift from Dr. Istvan Ando). Incubations with Alexa fluor conjugated secondary antibodies (1:1000 from Molecular Probes) were carried out at room temperature for two hours. Subsequently samples were washed in PBST and mounted in VECTASHIELD.
2.5. Cell proliferation assay in ex vivo cultured organs
Cell proliferation assays were performed in ex-vivo cultured organs by incorporation of 5-ethynyl-2′-deoxyuridine (EdU) using the Click-iT® Plus EdU Alexa Fluor® 555 Imaging Kit (Cat. No. C10638, ThermoFisher Scientific). For ex vivo culture, wandering third instar larvae were surface sterilized by briefly washing in 70% ethanol and rinsing in sterile PBS. The cleaned larvae were “inside out” dissected in optimized wing disc culture medium (WM1)(Zartman et al., 2013). Organs, including the brain and the imaginal discs that were present in the anterior half of the larvae, were exposed outside, but kept attached to the cuticle with tracheal and neuronal connections. The gut was removed to reduce bacterial contamination. The live tissues were gently washed in media several times and finally were placed in fresh medium for growth. For cell proliferation assays, the media contained EdU (10 μg/mL final concentration) from the Click-iT® Plus EdU Alexa Fluor® 555 Imaging Kit. The final volume of the media was optimized to maintain proper oxygen supply in tissues. The organs were incubated for 14 hours at 25°C. After incubation, media containing EdU was removed and the tissues were fixed in 4% PFA. These tissues were then washed twice with 1 mL 1 × PBS. The cells were then permeabilized by incubating the tissues in PBST (PBS + 0.1% Triton X-100) for 20 min at RT. Reaction cocktails, including 1X Click-iT® reaction buffer (880 μl), Copper protectant (20 μl), Alexa Fluor® picolyl azide (2.5μL) and 1X reaction buffer additive (100 μl), were prepared following the manufacturer's protocol. Finally, tissues were washed twice in PBS and incubated in 1 mL Click-iT reaction cocktail for 30 min in the dark with gentle rocking. After incubation, the organs were washed 3 times with PBS and the organs were dissected out and mounted on slides.
2.6. Hypoxia induction in ex vivo cultured organs
Drosophila organs were cultured ex vivo as described above. The organs were grown in fresh culture media either in presence or absence of CoCl2 for about 18 hrs in 25°C. Tissues were fixed, washed and mounted in VECTASHIELD for subsequent imaging.
2.7. RNA isolation and RT-PCR
Total RNA was purified from five adults of bnl-LexA/TM6, wild type (white), and nos-Cas9 parent flies using TRI Reagent (Sigma-Aldrich) followed by Direct-zol RNA purification kits (Zymo Research). To verify transcription of a hybrid lexA-bnl mRNA, RT-PCR was carried out from the total RNA by using primers RT-r and RT-f (Figure 2; # 15 and #16 in Table 2) to amplify an expected product of 433 bp by using One Taq one-step RT-PCR kit (NEB).
2.8. RNA in-situ hybridization
RNA in situ hybridization for bnl was carried out as described earlier (Guha et al, 2009) with few modifications. Embryos were fixed in 8% paraformaldehyde for in situ hybridization. A DIG labeled antisense riboprobe was generated by T3 RNA Polymerase (Promega)-mediated in vitro run-off transcription from a HindIII digested Bnl cDNA (pBSKII-Bnl). To detect the DIG labeled probe in situ, alkaline phosphatase conjugated α-DIG antibody (1:2000, Roche) treatment followed by NBT/BCIP (Roche) staining were used.
3. Results
3.1. Limitations of bnl-Gal4 drivers in reporting reliable bnl expression
Most studies rely upon a bnl-GAL4 driver (NP2211) for expression of transgenes in bnl-expressing cells. Although this GAL4 line closely mimics the bnl expression pattern in larva and pupa (Chen and Krasnow, 2014; Roy et al., 2014; Sato and Kornberg, 2002), we found that the GAL4 insertion line is less than ideal as a reporter in embryos. Expression driven by bnl-GAL4/UAS failed to mark bnl sources around the tracheal placodes in embryonic stages, and showed an abnormal expression profile that was inconsistent with bnl RNA in situ hybridization patterns (Sutherland et al., 1996) and bnl-lacZ enhancer trap expression (Fig 1A-B″). Similarly, the mCD8:GFP reporter driven by bnl-IT. Gal4 (InSight Target-Gal4, inserted at the 2nd intron of bnl) (Gohl et al., 2011) did not mark the bnl expressing cells in embryo or in larva (Figure 1C-C″, F). Therefore, to extend the genetic tool box for expression of transgenes specifically only in the bnl sources, we aimed to generate a LexA/lexO-based binary transcription system. Due to the unreliable expression patterns of bnl-GAL4 or bnl-IT.GAL4, repurposing these lines into the QF or LexA based systems using homology assisted CRISPR knock-in (HACK) (Lin and Potter, 2016) or Integrase Swappable In Vivo Targeting Element (InSITE) (Gohl et al., 2011) was unsuitable.
3.2. Generating a bnl-LexA driver with CRISPR/Cas9 based genome editing
The cis-regulatory elements (CREs) that drive complex spatiotemporal bnl expression are not known. Therefore, an efficient strategy to drive expression of LexA in an accurate pattern of the endogenous bnl gene can be achieved by the genomic replacement of the first exon of bnl with the sequence of the LexA transactivator. For replacing the bnl exon with the LexA sequence, we selected an improved nls-LexA∷p65 cassette that was shown to provide optimal expression in Drosophila (Pfeiffer et al., 2010). However, it is difficult to precisely delete an 835 bp large defined genomic region and replace it with ∼1.8kb long DNA fragment containing the LexA cassette. There is no precedence for such exon-swap by CRISPR/Cas9 based genome editing. In addition, the bnl mRNA has a long 5′ UTR (Fig 2A) and is known to localize in a subcellular compartment (Jarecki et al., 1999). Dynamic bnl expression can also be post-transcriptionally controlled at the levels of mRNA maturation, localization or translation. Therefore, we incorporated several unique strategies to generate bnl-LexA.
To preserve most transcriptional and post-transcriptional regulations of the bnl gene on the LexA reporter expression, about 835 bp in the middle region of the first bnl coding exon were targeted to be replaced by the nls-lexA:p65 coding sequence. This strategy aimed to generate a chimeric nls-lexA:p65-bnl mRNA, where the original long bnl 5′ UTR, a 93 nucleotide region (first 31 amino acids of N-terminal part of Bnl protein) of the 5′ end, and a 48 nucleotide region of the 3′ end of the edited coding exon along with the complete 3′ bnl gene sequence were retained in the engineered lexA mRNA product. Thus, the chimeric lexA mRNA would be synthesized under bnl transcriptional control and it would retain all original bnl exon splicing, post-transcriptional and translation controls. To avoid translation of LexA:p65 as a chimeric Bnl protein, a T2A self-cleaving peptide was inserted between the N terminal 31 amino acids of the bnl coding region and the start codon of nls-lexA:p65 (Fig. 2A). T2A facilitates “ribosome skipping” (González et al., 2011) and independent translation initiation of LexA. Similarly, to disrupt co-translation of a truncated C terminal portion of Bnl from the hybrid transcript, the nls-lexA:p65 is followed by a stop codon. Thus, in the genome edited fly, a completely functional LexA protein was expected to be produced from the engineered mRNA. A replacement cassette containing all these features was generated (Materials and Methods).
To efficiently delete a large, defined region of the exon, we used two separate gRNAs that specifically targeted only the two ends of the defined region in the bnl locus. These gRNAs, along with the replacement cassette, were co-injected and expressed in the embryonic germline to induce Cas9 dependent DSB and replacement (Fig. 2A). Subsequently, the progenies were screened for the intended replacement as described earlier (Materials and Methods). The genome editing process generated progenies that were randomly derived either by ends-out recombination with the intended replacement, or alternatively, by unintended ends-in recombination. In the ends-in recombination product, the replacement vector backbone is normally inserted in the genome along with a duplication of the targeted region. About 67% of injected G0 animals were found to be fertile. Using a three step PCR based screening approach (see Materials and Methods, Fig 2B, C, Table 1) we found that within the fertile G0 lines, ∼56% were the founders, which transmitted HDR to the next generation. About 23% of tested F1 progeny were positive for successful HDR, and ∼73% of positive lines were “ends-out” HDR. Compared with previous reports of approximately 18% germline transmission rate using two gRNAs for targeted replacement (Gratz et al., 2014; Li et al., 2015), a much higher efficiency was obtained in this procedure. This is probably due to improved injection efficiency or optimal activities of the selected gRNAs. Since the first coding exon of bnl is replaced by the lexA, all the HDR lines are homozygous lethal and are maintained over balancers.
Table 1. Efficiency of CRISPR/Cas9 mediated HDR.
| Male crosses | Female crosses | Total | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| # founders | # HDR progeny | # founders | # HDR progeny | # germline transmission | # HDR progeny | # “ends-out” HDR |
| 7/15 | 37/150 | 8/12 | 26/120 | 15/27 | 63/270 | 46/63 |
Number of corresponding genome edited flies out of total is shown in each column.
To determine whether the genome edited bnl-LexA lines are expressed in the same pattern as the endogenous bnl, each positive HDR line was crossed to lexO-mCherryCAAX transgenic flies. The progeny embryos and larvae were examined for the reporter expression pattern. Out of 50 lines that were verified, 42 showed expression consistent with the previously characterized bnl patterns in embryo and larval wing imaginal discs. Synthesis of the chimeric mRNA containing the sequence of bnl91nt-T2A-nls-lexA:p65-translation stop-bnlc in the genome edited fly was confirmed by RT-PCR analysis on total RNA from several lines (Figs. 2, S1A, Materials and Methods). This hybrid bnl mRNA product can only be synthesized from the endogenous bnl transcriptional and post-transcriptional control in the genome edited progenies. These results suggested a successful application of the strategies used in this study. Two completely sequence-verified “ends-out” lines were used in the subsequent studies to thoroughly analyze the specificity and versatility of the bnl-LexA.
3.3. Expression of bnl-LexA in previously characterized bnl patterns
Expression of the bnl-LexA was verified in embryonic, larval and adult tissues by simultaneously expressing CherryCAAX (lexO-mCherryCAAX) in the bnl source (bnl-LexA) and CD8:GFP (UAS-CD8:GFP) in the tracheal cells (btl-GAL4). Accurate expression of bnl-LexA was confirmed in the wing imaginal discs, Tr4 and Tr5 transverse connective cells, dorsal branch of the Tr2 tracheal metamere, and in male genital disc cells. In these tissues the role of Bnl signaling had been thoroughly characterized (Sato and Kornberg, 2002; Chen and Krasnow, 2014; Ahmad and Baker, 2002) (Fig 3A-D). Consistent with previous reports (Jarecki et al., 1999), bnl-LexA was also detected to be broadly expressed in most larval tissues (Fig. S1C). The LexA driver did not express in non-specific locations as the bnl-Gal4 line, which non-specifically expressed in some larval tissues including in salivary gland, an organ devoid of bnl expression (Fig. S1C). Expression of bnl-LexA in various stages of the embryo (Fig 3E-J) were consistent with a previous report (Sutherland et al., 1996), RNA in situ hybridization, and the bnl-lacZ patterns (Figs. 1, S2).
Figure 3. Expression of bnl-LexA in developmental stages and in hypoxia. A-A′.
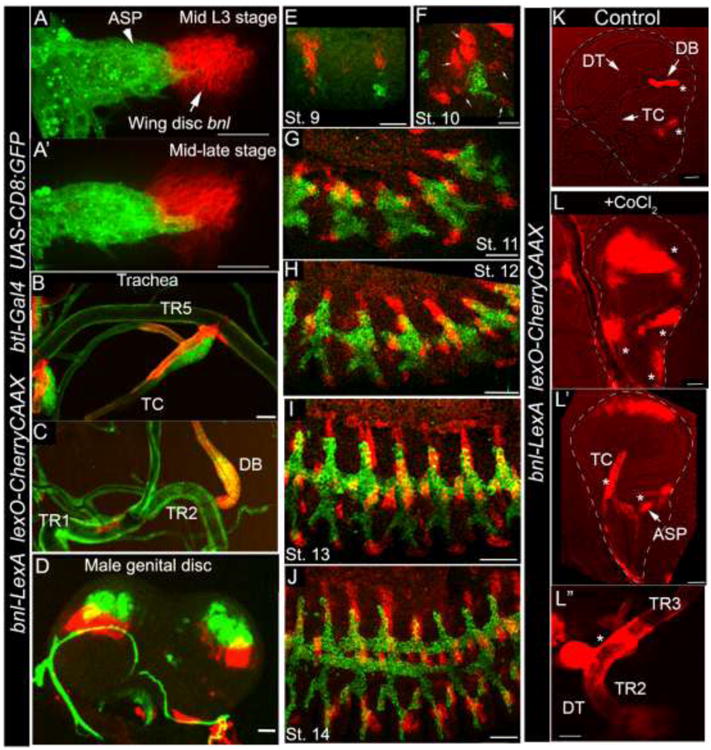
Wing disc bnl source (red) and the ASP (green) in mid and late L3 larval wing discs. B-C; bnl expression in larval tracheal branches: TR5 transverse connective (TC) (B); TR2 dorsal branch (DB) (C). D, bnl expression (red) in genital discs. Mesenchymal cells associated with the bnl expressing disc epithelium express btl (green). E-J, confocal images of fixed embryos of different stages; Small arrows, five bnl sources surrounding the tracheal placode. Genotypes: A-J; btl-Gal4, UAS-CD8:GFP/+; bnl-LexA,lexO-herryCAAX/+. K-Lhypoxia induced bnl-LexA expression profile in wing discs and associated TR2 tracheal metamere (TC, DB, dorsal trunk-DT). Genotype: bnl-LexAlexO-CherryCAAX/+. K, control discs from ex vivo cultured organs without CoCl2. L-Lwing discs (L, L′) and trachea (DT, (L″)) from cultured ex vivo organs with CoCl2 induced hypoxia. Star, ectopic expression. K-L″; Images were captured with 555 nm fluorescence as well as with bright field. The superimposed images were shown. Scale bars, 30 μm; K-L″, 50 μm.
Expression of bnl is induced in hypoxia by the oxygen-regulated a subunit of Hypoxia-Inducible Factor (HIF), Similar (Sima, HIF1a) (Jarecki et al., 1999; Centanin et al., 2008). To verify hypoxia induced response of bnl-LexA, larval organs were cultured ex vivo in presence or absence of CoCl2 for 14-18 hours (Materials and Methods). CoCl2 is a chemical inducer of HIF1 α (Triantafyllou et al., 2006; Wu and Yotnda, 2011). Cultured organs that were grown in presence of CoCl2 induced bnl-LexA expression in ectopic locations such as in the wing disc pouch, tracheal cells in the TR2 branches and ASP (Fig. 3L-L″). Expression of bnl in these ectopic locations was not observed in tissues that were cultured without CoCl2 (Fig. 3K). Thus, bnl-LexA expression can be conditionally induced under hypoxia. These results confirmed that bnl-LexA is a reliable tissue specific bnl reporter driven by genomic bnl CREs in all developmental and metabolic conditions. Thus, this genetic tool can reliably be utilized to explore bnl expression in various tissues and study live cellular interactions.
3.4. Morphogenetic movement of embryonic bnl-LexA expressing cells
The current model of tracheal branching morphogenesis proposes that bnl is dynamically expressed in epidermal cells ahead of the growing tracheal branches, to guide them to their destination. In concordance with this model, images of the fixed or live embryos, which expressed CherryCAAX in the bnl source and CD8:GFP in the trachea, detected an induction of bnl expression in five epidermal clusters around the tracheal placode between stages 9 and 10 (Fig. 3E,F). Live four-dimensional imaging, focusing on the dorsolateral bnl-expressing clusters around the tracheal placode, detected a temporal increase of expression level in the five clusters after stage 11 (Fig 4A, Supplementary movies 1-9). However, unlike predicted by the model, a dynamic change in location of bnl expression in the new epideramal cells ahead of the growing dorsal branch was not detected. Instead of the epidermal cells surrounding the growing dorsal branch, many amnioserosa cells activated bnl expression from stage 13 onwards (Figs. 4A; Supplementary movies 1-9; S2B,B′). Toward the end of embryogenesis, bnl expression was also induced in additional tissues, especially in muscles (Fig 4H-J, supplementary movies 3-5). During dorsal closure, amnioserosa cells delaminated out of the dorsal part (Fig 4A; supplementary movies 1, 3, 4). Although the significance of bnl expression in amnioserosa is not clear, a late stage induction of bnl expression in muscles may regulate the growth of tracheal branching into the tissue at a subsequent developmental stage. Thus, from these results it can be predicted that the spatiotemporal activation of bnl expression is a major stage-specific event that is necessary to signal when and where a tracheal branch would grow. However, the dynamic spatial change in bnl expressing epidermal cells ahead of a growing tracheal branch did not appear to be due to the rapid switching on or off the transcription of the gene.
Figure 4. Migration of bnl-LexA expressing cells in embryonic development. A-J.
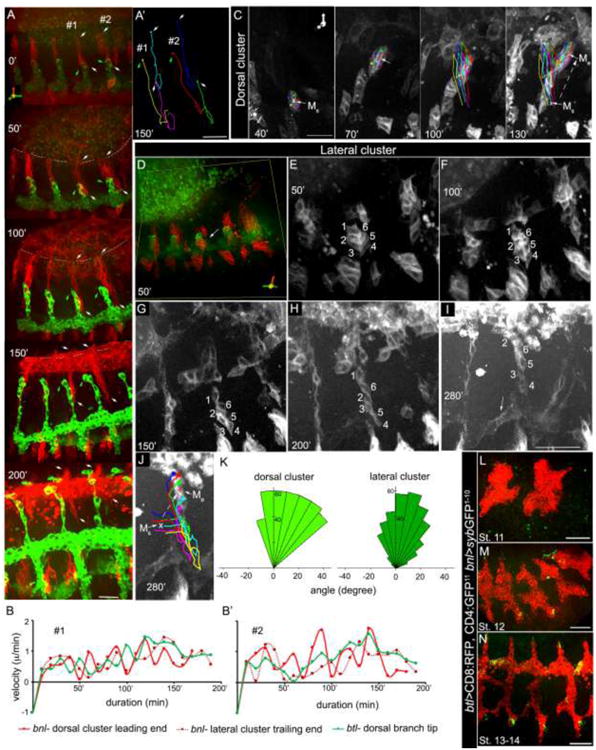
morphogenetic movement of dorsolateral bnl-clusters and corresponding dorsal branch (green) in each body segment; live time lapse images taken once every 10-min duration. A, 3D projections of time lapse images at 0 (stage 12), 50, 100,150 and 200 min of a growing embryo; intimate interactions and migrations of both dorso-lateral bnl-clusters (red) and dorsal branch (green) were detected; epithelial edge in dorsal closure is marked by dashed line; #1-2, two body segments; downward arrows (white), leading edges of dorsal clusters in #1 and #2; upward white arrows, trailing ends of the lateral clusters in #1 and #2; green arrows, tips of the dorsal branches in #1 and #2; converging tracks of dorsal bnl-clusters over the amnioserosa were visible at 200 min; induction of bnl expression in amnioserosa was visible from 50 min onwards; a gradual increase in bnl transcription in dorsolateral clusters was detected from 0-150 min; see corresponding Supplementary Movie-1. A′ tracked migration routes (from 0-150′) of the -leading ends of dorsal bnl-clusters, -trailing ends of the lateral bnl-clusters, and the corresponding tracheal tips in #1 and #2 segments of the embryo shown in (A). B, B′; coordinated growth kinetics of the leading edge of the dorsal bnl-cluster (red), trailing end of the lateral bnl- cluster (purple dash line) and corresponding tip of dorsal branch (green) in #1 (B) and #2 (B′) body segments of the embryo shown in (A); see corresponding Supplementary Movies 4, 9. C, tracked migration routes of six cells constituting a dorsal cluster in 3D space from 40 min (stage 12) to 130 min of imaging; panels showing images at every 30 min duration; Ms, center of mass (white x) of the group at the origin; Me, center of mass (red +) of the same group at the end of experiment; dashed arrow shows the displacement of the center of mass of the dorsal cluster in 2D; see corresponding movies 3,5,7. D-J, showing a lateral bnl-cluster from a posterior segment of an embryo (D) containing six cells and their relative position in the cluster at various time point (E-J); see corresponding supplementary movies 2, 6; J, migration tracks of the six cells in lateral cluster from 30 to 280 min of recording (see Supplementary movie 8); white dashed arrow, the displacement of center of mass in 2D. K, rose plots showing directed migration of each of the six cell in a dorsal cluster (left) and in a lateral cluster (right) over time from posterior segments of two different embryos that are similarly oriented while imaging; plots show a coherent dorsal-ward migration. L-N, patterns of cellular contacts between the tracheal tip cells and the bnl source marked by syb-GFP reconstitution at different embryonic developmental stages; bnl-expressing cells expressed nsyb-GFP1-10 and trachea expressed CD4:GFP11 and CD8:RFP (btl-Gal4; bnl-LexA X lexO-nsybGFP1-10, UAS-CD4:GFP11; UAS-CD8:RFP); All embryo panels, anterior - left, dorsal - up. A-J, time durations are relative to the 0 min (start of imaging, normally stage 11-12); genotype: A-J, btl-Gal4, UAS-CD8:GFP; bnl-LexA, lexO-mCherryCAAX. Scale bars, 30μm.
Instead of transcriptional activation, live time-lapse imaging of embryos revealed a remarkable dorsolateral reorganization and morphogenetic movement of bnl-expressing cells that coincided with the tracheal branching processes (Fig 4A-J, Supplementary movies 1-9). In each body segment the lateral cluster merged with the dorsal cluster and collectively migrated dorsal-ward (Fig. 4A, E-J; Supplementary movies 4,5,6). To accurately determine the movement of bnl-clusters, the leading and trailing ends of the dorsolateral clusters, as well as the corresponding tips of the dorsal tracheal branches were tracked at every 10-minute duration from the developing embryos (n=6, Fig. 4A-A′; Supplementary movies 4,9). These tracking experiments revealed several important features of migration of the epidermal bnl-source.
(1) In each body segment, cells in the dorsal clusters converged from the two opposing sides into the dorsal midline and moved on top of the bnl-expressing amnioserosa cells. The leading edge of the dorsal cluster moved at an average rate of ∼ 0.6 μm/min (measured from multiple embryos) and appeared to constitute the converging epidermal edges of the dorsal closure (Fig 4A; Supplementary movie 1, 3, 4, 5,9). This migration rate is similar to the rate of dorsal closure (Haack et al., 2014). Although at this time the migratory front of the dorsolateral clusters could not be tracked due to high level of CherryCAAX expression in the amnioserosa, each segmental track, constituted of bnl expressing cells, was visible over the amnioserosa (Fig. 4A). Coordination of dorsal-ward migration of the bnl-expressing cells with the dorsal closure may indicate that the movement of the bnl-expressing cells may be a part of the global morphogenetic cell movements that take place in gastrulating embryo.
(2) The average rate of migration of the tracheal tips was ∼ 0.7 nm/min. By plotting the migration rates of the leading edge and trailing ends of a dorsal and lateral bnl-clusters and the corresponding dorsal branch in a body segment, a rhythmic oscillating pattern of high and low activity for both trachea and bnl source was observed (Fig 4B, B′). The oscillating phases of migration in these two tissues appeared to be slightly shifted during early stages, but synchronized in later stages of development. The rate of migration of bnl-source and trachea varied slightly among the middle and the posterior body segments of an embryo or among different embryos. However, the pattern of oscillating phases of migration rates remained consistent for all individual dorsal branches and their corresponding dorsolateral bnl- clusters, measured in the same or different embryos (Fig. 4 B, B′; not shown for other embryos). Therefore, the migration of the ligand expressing dorsolateral clusters surrounding a dorsal branch is closely coordinated with the migration of the dorsal branch.
(3) In all embryos, the leading cells of the lateral clusters were found to merge with the trailing ends of the dorsal clusters, contact the tracheal tips and collectively migrate dorsal-ward (Fig. 4A,C, E-I; Supplementary movies 1,5,6,9). The migration routes of each of the cells in dorsal and lateral clusters are highly coherent and polarized toward the dorsal midline (Fig 4K). Together, the cells in dorsolateral clusters in each segment appeared to form a motile elongated track of bnl source ahead and under the growing dorsal branch.
(4) To further examine whether the same group of bnl-expressing cells collectively migrate, individual bnl-expressing cells were tracked over time from multiple dorsal and lateral clusters in several developing embryos. The clusters that were in clear focus in 3D projections were tracked and were consistently found to contain ∼six cells since late stages 11-12 (Fig 4C, D-I, supplementary movies 5-8). The same group of cells reorganized their positions, changed their shapes to render an elongated morphology to the cluster, and collectively migrated toward the dorsal mid line. Thus, the results here suggested a model where the morphogenetic movement of the bnl-clusters creates the dynamic spatiotemporal pattern of signal source ahead of a growing tracheal branch to precisely regulate its directional migration.
A close proximity of each of the bnl-expressing clusters with the primary tracheal branches was noticed at all times during development (Fig 4A,D). In live cell imaging (Supplementary movies 1, 2), the migrating cells of the dorsal tracheal branch and the corresponding dorsolateral bnl-expressing clusters were found to establish direct contacts for a number of times. The qualitative pattern of these contacts was revealed by employing a synaptobrevin-GFP Reconstitution Across Synaptic Partner (syb-GRASP) (Macpherson et al., 2015) technique (Materials and Methods). In this experiment, only the tips of the growing primary branches were found to establish the most stable contacts with the bnl sources. The patterns of contact changed with the developmental stages of the embryo (4L-N). These results showed that although a growing tracheal branch and bnl-expressing cells move in close apposition with one another, most of the contact dependent communication between these two tissues takes place around the growing tracheal tips.
3.5. bnl-LexA expression identified a tracheoblast niche in leg imaginal discs
Exploration of the bnl-LexA patterns in various larval tissues identified a new bnl-source in all three pairs of the L3 larval leg imaginal discs. A btl-expressing tracheal tube was found to be associated with the bnl-source (Fig. 5A-D). Immunostaining for bnl-lacZ and RNA in situ hybridization confirmed the specificity of this expression pattern (Fig S3). In leg discs, bnl-LexA marked a narrow band of about 30-40 epithelial cells along the anterior/posterior axes of the notum and the stalk region of the discs (Figs. 5A-B′, S3A). The epithelial nature of the bnl-expressing cells located in the notum was depicted from the E-cadherin localization in the cell junctions (Fig 5C,C). The tracheal tube associated with the Bnl source contained about 30-40 btl expressing cells in leg disc notum. Apical E-Cad localization clearly marked the lumen of the newly growing tube. In the stalk region of the leg disc, this tracheal tube was found to be wrapped around by the bnl-expressing disc epithelial cells (Fig. 5B,B′; supplementary movie 10). EdU incorporation experiments suggested that the new tracheal branch contains proliferating tracheoblasts (Fig 5E, supplementary movie 11; see Materials and Methods).
Figure 5. Expression of bnl-LexA in tracheoblast niche of the leg imaginal discs. A-B′.
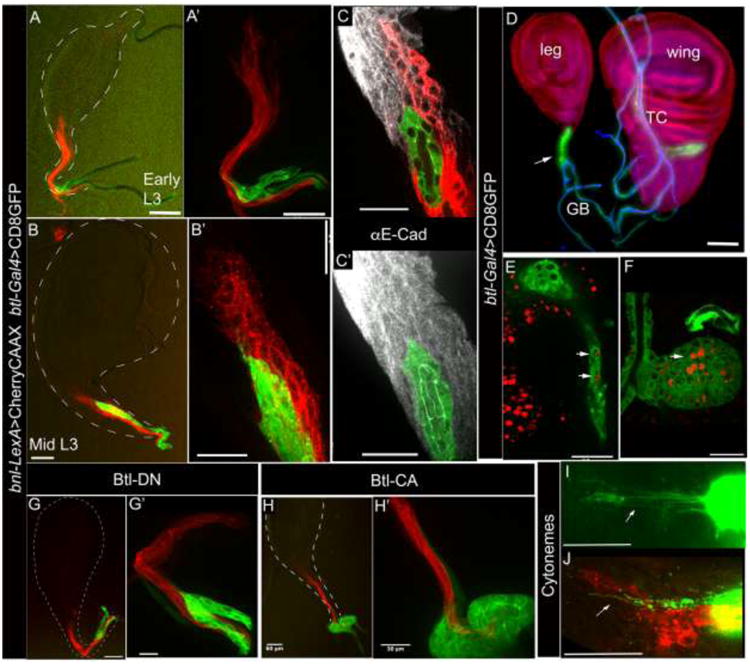
bnl-LexA expression pattern in early (A-A′) and mid (B-B′) developmental stages in the L3 larval leg discs (3rd leg). A′, B′; 40X images of A and B, respectively. C, C; Anti-E-Cad staining marked cell junctions of the disc epithelial bnl source (red; C) and the apical (green) lumen (C) of the tracheal tube lined by the tracheoblast cells. D, wing- and 3rd leg imaginal discs with associated neo-tracheal tubes containing tracheal progenitors (btl-Gal4 X UAS-CD8:GFP; arrow); ASP originates from the TR2 transverse connective (TC) in wing disc and leg disc tracheoblasts migrate from the ganglionic branch. Cell nuclei marked by Histone-RFP (His2Av-mRFP1) transgene. Chitinous lumen of the tracheal tubes (blue) were imaged with autofluorescence in 405 nm channel. E, F; cell proliferation detected by EdU incorporation (red) in ex vivo cultured wing and leg discs and in the associated tracheal progenitor cells in the leg- (E, see supplementary movie 11) and in the wing- discs (F). G-G′; Btl-DN expression in trachea suppressed tracheal migration into the leg disc but did not affect disc bnl expression (btl-Gal4, UAS-CD8:GFP/+; bnl-LexA,lexO-CherryCAAX/UAS-btlDN); G′, 40X image of G. H-H′; Activated-Btl expression (btl-Gal4, UAS-CD8:GFP/+; bnl-LexA,lexO-CherryCAAX/UAS-ABtl) in trachea resulted in abnormal growth of tracheoblast cells at the base of the leg disc but these cells did not follow the bnl -expressing cellular track into the leg discs; H′, 40X image of H. I-J, Cytonemes (arrows) at the tip of the growing tracheal branch in leg disc in live (I) and in fixed condition (J). Genotypes: A-C, J; btl-Gal4,UAS-CD8:GFP/+; bnl-LexA,lexO-CherryCAAX/+. E,F,I; btl-Gal4, UAS-CD8:GFP. Scale bars, 30 μm; A,B,D,G,H, 60 μm.
The second-instar larval leg-discs expressed bnl-LexA, but did not contain the tracheal tube. At this stage, the btl-expression was detected only in the secondary tracheal branches that are attached to the base of the leg disc stalks (Fig. 5A, A′). Thus, the tracheoblast cells from these secondary branches appeared to follow the track of bnl-expression to migrate into the leg discs only during the L3 larval stage (Fig. 5D, S3F). The secondary branch for the first pair of leg discs was found to be the cerebropharyngeal branch (Manning and Krasnow, 1993) from TR1 (Fig. S3I). The second and third pairs of leg discs are connected by the ganglionic branches from the TR2 transverse connective (Fig. 5D, S3H). To verify whether Bnl signaling may control the migration of these tracheoblasts into the leg disc, either the dominant-negative (Btl-DN) or the activated Btl (λBtl) was expressed in the tracheal cells. The Btl-DN expressing tracheoblast cells did not migrate into the leg discs as they were incapable of activating Bnl signaling (Fig 5G, G′). Ligand-independent signaling activation by expressing λBtl (Sato and Kornberg, 2002) in trachea increased cell-autonomous proliferation. An increased number of tracheal cells accumulated at the base of the leg discs and did not migrate into the disc following the bnl-expressing cellular track (Fig. 5H, H′). Thus, bnl-expressing cells in the leg discs control directional migration and morphogenesis of the newly identified tracheoblast niche. Interestingly, similar to ASP cells, the leg disc tracheoblasts were found to extend signaling filopodia or cytonemes toward the bnl-expressing cells, probably to receive Bnl (Roy and Kornberg, 2015)(Fig. 5I, J).
3.6. Neuronal bnl and btl expression in larval eye discs
An earlier report (Jarecki et al., 1999) showed that bnl is expressed in the eye imaginal disc epithelium, but the disc is devoid of tracheal growth. This result suggested a non-tracheogenic role of bnl in larval eye disc. Simultaneous expression of CherryCAAX from bnl-LexA and CD8GFP from btl-Gal4 in larval eye disc revealed that posterior to the morphogenetic furrow, both the btl- and bnl-expressing cells represented Elav positive neurons of the developing retina (Figs. 6A,B; S4D,E). The bnl expression pattern was confirmed with bnl in situ hybridization, bnl-Gal4 and bnl-lacZ patterns (Fig. S4A-C). The bnl expressing neurons were recruited only to a subset of ommatidial clusters that were localized along the dorsal and ventral most edges of the disc, converging into the optic stalk (Fig. 6A-C). Since bnl is expressed in both retinal neurons as well as in the dorsal and ventral corner of the undifferentiated epithelia anterior to the furrow (Figs. 6A; S4 A-C), it is likely that bnl expression is not specifically induced in the ommatidial clusters. Rather, these neurons are probably specified from the bnl- expressing epithelial cells situated along the edges of the disc during the progression of MF. In each ommatidium, the bnl-expressing neurons were the Senseless positive R8 photoreceptors (Fig. 6D). In contrast to bnl expression, btl-expressing cells were localized only in the differentiated retina (Figs. 6A; S4B). They were found to be Prospero positive R7 photoreceptors (Figs. 6E). All bnl-expressing ommatidial clusters contained btl-expressing R7 cells. Although some of the ommatidial clusters contained only the btl-expressing R7 cells without detectable bnl expression in them, axonal projections from all the btl-expressing R7 and bnl-expressing R8 neurons were in close apposition in the optic stalk and fasciculate together into the optic lobe (Figs. 6A, E-H; S4 B,F; Supplementary movies 12, 13).
Figure 6. Neuronal expression of bnl in larval and embryonic development. A.
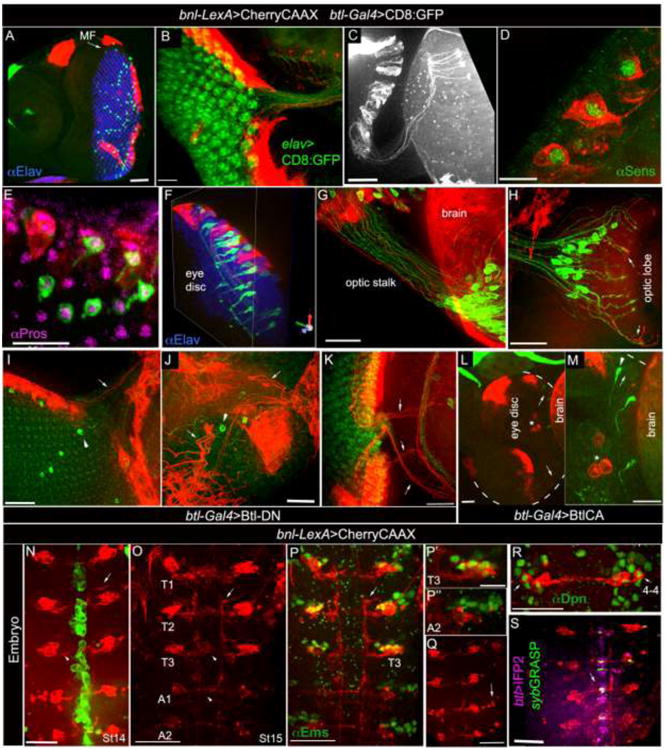
bnl-LexA and btl-Gal4 expression in larval eye disc; Elav immunostaining (blue) marked retinal neurons posterior to the morphogenetic furrow (MF). B, bnl-LexA expression marked neurons in ommatidial clusters; elav-Gal4 driven CD8:GFP marked all neurons (UAS-CD8:GFP X elav-Gal4). C, ommatidial bnl-neuron extended axons through optic stalk into the optic lobe. D, 3D projection showing bnl-expressing neurons marked by Sens immunostaining, a marker for the R8 photoreceptors. E, 3D projection showing Pros immunostaining that marked btl-expressing neurons; Pros, R7 photoreceptor marker. F, 3-D projection showing intimate organization of btl- (green) and bnl- (red) expressing neurons in the eye disc ommatidia (also see supplementary movies 12,13); F-H, axons from receptor (Btl) and ligand (Bnl) expressing neurons fasciculate through optic stalk into the optic lobe; arrow, fasciculation in optic lobe. I-M, phenotypic consequences in bnl- and btl- expressing neurons when Btl-DN (I-K) and λBtl (L-M) were expressed specifically in the btl-neurons; arrow, bnl-expressing neurons; arrowhead, btl-expressing neurons; star, hemocytes; dashed line (L-M), eye disc out line; L, M, same sample in two different magnifications. Genotypes: A, E- H, btl-Gal4, UAS-CD8:GFP/+; bnl-LexA,lexO-CherryCAAX/+; C, D, bnl-LexAlexO-CherryCAAX/+; I-K, btl-Gal4, UAS-CD8:GFP/+; bnl-LexA,lexO-CherryCAAX/UAS-Btl-DN; L, M, btl-Gal4, UAS-CD8:GFP/UAS-XBtl; bnl-LexA, lexO-CherryCAAX/+. A-H, eye disc anterior-left. N-S, stage 14-16 embryos, ventral view, anterior up. N-O, bnl-LexA expression in neurons (red) and btl-Gal4 (N) in the ventral midline (green) of stage 14 (N) and stage 15 (O) embryos; bnl-LexA expressing neurons in thoracic segments (T1-T3), had both interneuronal (arrowhead) and motor neuronal (arrow) projections, and had only interneuronal projections in abdominal hemisegments. P-P″, immunostaining for Ems, a marker for NB 3-5 and 4-4 lineages, marked bnl-expressing neurons in T1-T3, but did not mark the abdominal ones; P′,P″, magnified view of Ems immunostaining from T3 (P) and A2 clusters (P″) shown in P. Q; punctate lobular fluorescence (arrow) next to the large cells bodies of the abdominal bnl-expressing NB lineage suggested apoptosis in several abdominal neurons. R, one cell (arrow) in each bnl-cluster is immunostained for Dpn, a neuroblast marker. S, synaptic contacts (green fluorescence) between btl-expressing cells (purple) and bnl-neurons (red), mapped (arrow) in the ventral midline by GFP reconstitution using nsyb-GRASP technique; genotype: btl-Gal4/ lexO-nsybGFP1-10, UAS-CD4:GFP11; bnl-lexA, lexO-CherryCAAX/UAS-CD4:IFP2. Scale bars, 30 μm; A, 60 μm; D, 10 μm.
To examine whether the close apposition of bnl- and btl- expressing neurons depends on Bnl-signaling, a dominant-negative form of the receptor (Btl-DN) was expressed in btl-expressing neurons using btl-Gal4. In this condition, only a few btl-neurons were found to be localized in the eye disc and in the brain lobe, but bnl-expression in the retina was unaffected (Figs 6I-K). The optic stalks were stunted and the axon terminals in the optic lobe were miss-targeted. Ligand independent activation of Bnl-signaling by overexpressing λBtl using btl-Gal4 led to a reduction in the number of both btl- and bnl-positive neurons in the retina (Fig. 6L-M; S4G). Concomitant with the reduction in bnl-expressing neurons, an increase in CherryCAAX marked eye disc associated hemocytes (marked by Hemese antibody, see Materials and Methods, not shown) was observed. This result suggested that ectopic activation of RTK signaling in neurons may lead to neuronal cell death. However, it is not clear whether the marking of hemocytes is due to an induction of bnl-expression in these cells or due to phagocytosis of the dying CherryCAAX marked neurons. In contrast to reduction in btl-positive neurons, Btl-CA mediated ectopic activation of signaling led to over-proliferation and ectopic branching of tracheal cells (not shown). Unlike the neuronal phenotype, epithelial bnl expression in the eye discs remained unaffected by Btl-CA expression in btl-positive cells (Fig. 6L). Taken together, these results may suggest that Bnl-signaling is important for the organization and maintenance of the visual circuit.
3.7. bnl-LexA expression in embryonic central nervous system
In the larval brain a Bnl and Hedgehog (Hh) feed-back loop regulates the onset of neuroblast division (Barrett et al., 2008). Although establishment of the Bnl-Hh feedback loop takes place in embryo, the identity of the embryonic bnl-expressing neuroblasts was not known. CherryCAAX reporter driven by bnl-LexA identified a segmental distribution of bnl-expressing neurons in the embryonic ventral midline (stage 13-16 embryo) (Fig. 6N-O). Elav positive (Fig S4I) neuronal expression of bnl was confirmed by immunostaining of bnl-lacZ and RNA in situ hybridization (Fig. S4H,J), but subcellular localization of the signal detected by these methods did not reveal a neuronal morphology. Engrailed, a marker for NB 6 and 7(Broadus et al., 1995), Hb9 (Extra-extra), a marker for NB 1-1, 1-2, 2-2, 3-1, 3-2, and 5-2 (Lacin and Truman, 2016), and the eagle-Gal4 line that marks NB 2-4 and 3-3 (Higashijima et al., 1996) did not localize in bnl-neurons (Fig. S4K-M). Immunostaining for Dpn (deadpan) identified a single NB situated closest to the epidermis in each of the bnl-expressing clusters (Fig. 6R). Considering all these expression patterns and the stereotyped arrangement of embryonic NBs (Doe, 1992), bnl-expressing neurons were predicted to be either from the NB 4-4 or NB 3-5 lineage. Positive immunostaining for Ems (empty spiracles), a marker for NB 3-5, and 4-4 (Lacin and Truman, 2016) confirmed that the bnl-expressing neurons are either NB 3-5 or 4-4 (Fig. 6P-P″). We reasoned that bnl-neurons are derived from NB 4-4 rather than NB 3-5 because similar to NB 4-4, bnl-expressing neuroblasts had relatively smaller nuclei in comparison to NB 3-5 (Fig. 6R). Secondly, the late-stage (stage 13) expression of bnl in neurons matched the timing of delamination of NB 4-4 (S4), rather than NB 3-5 (S1). Finally, similar to the NB 4-4 lineage (Lacin and Truman, 2016), the bnl-expressing neurons were found in all thoracic and abdominal segments, both had interneuron and motor neuron projections in the thoracic segments, and they induced apoptosis toward late embryogenesis and were not marked by Ems in the abdominal hemisegments (Fig. 6O-R).
Interestingly, Btl is known to be expressed in the embryonic midline cells, which include glia and Ventral Unpaired Median (VUM) neurons (Klämbt et al., 1992) (Figs. 6N,S, S4J). It is possible that the bnl-expressing NB lineage may provide the signaling cue for these Btl expressing midline cells. To visualize the direct synaptic contact between the bnl-expressing neurons with the btl-positive midline cells, the sybGRASP technique was employed (see Materials and Methods). The bnl- and btl-expressing neurons were separately marked by CherryCAAX and CD4:IFP2 (Infra-red fluorescent protein, (Yu et al., 2014)), and they expressed either sybGFP1-10 or CD4:GFP11. The active synaptic sites between the receptor and ligand expressing cells were detected by reconstituted GFP signal in the ventral midline (Fig 6S). These synaptic sites may represent the putative sites for Bnl signal exchange. Thus, this study identified the NB lineage of the bnl-expressing neurons and defined their organization and interaction with the receptor expressing midline cells in the embryonic CNS.
4. Discussion
In this study, we described an efficient strategy of exon replacement using CRISPR/Cas9 based genome editing and its application in generating a bnl-specific LexA/LexO based binary expression system that is under the same transcriptional and post-transcriptional control of the gene. This new genetic tool is the only available binary transcription driver found so far to be highly target specific and versatile for use in all developmental and metabolic conditions. Thus, the target specific bnl-LexA driver markedly expands the available genetic tool box to (i) conveniently and reliably analyze spatiotemporal expression patterns in live and fixed tissues, (ii) characterize lineage and functional organization of bnl expressing cells, and (iii) simultaneously label and manipulate gene activities in btl- and bnl-expressing cells to investigate the mechanism of signaling interaction between signal exchanging cells. Combining the reporter expression driven by bnl-LexA with btl-Gal4 lines we were able to characterize the unique spatiotemporal organization of several new bnl and btl- expressing cells, especially during tracheal and neuronal development.
Previous studies identifying tracheoblast cells in larval wing disc associated ASP (Sato and Kornberg, 2002) hinted at possible additional tracheoblast niches in other imaginal discs. By exploring the bnl-LexA expression patterns in different larval tissues, we identified a new bnl-expressing tracheoblast niche in leg-imaginal discs. Genetic manipulation experiments suggested that the Bnl signal produced from the leg disc cells acts as a local guidance cue for the directed migration of tracheoblast cells from the disc-associated secondary tracheal branches into the 3rd instar leg discs. The adult fate of the tracheoblast cells in the leg disc associated neo-tracheal branch could not be mapped. However, considering the organization and growth of this btl>GFP marked branch in the late L3 larval-, prepupal-, and pupal- leg discs, as well as the location of adult air sacs in pharate adults (Fig S3L-N), we speculate that the leg disc tracheoblasts may form the ventral air sacs (prepleural, sternopleural and hypopleural) and parts of the leg trachea (Bodenstein, 1965).
An interesting insight on how Bnl signaling may control tracheal branching processes has emerged from live imaging analyses of embryos. In contrary to the existing model of tracheogenesis, where spatiotemporal bnl pattern is predicted to be a result of dynamic regulation of bnl transcription, the current study suggested a role of morphogenetic movement of the bnl-expressing cells in creating the dynamic expression pattern (Fig 4, Supplementary movies 1-9). A dynamically moving signal source may spatiotemporally restrict the availability of the Bnl signal to a growing tracheal branch and can precisely regulate the directional migration of a growing branch. However, importance of a stage-dependent complex regulation of bnl CREs is also evident at an early stage activation of the gene in five epidermal clusters or in induction of bnl in the muscles during late embryogenesis. Thus, a combination of spatiotemporal transcription of bnl with the morphogenetic movement of bnl-expressing cells may be necessary to precisely regulate the timing and location of tracheal budding and subsequently to control the directional migration of the tracheal branch. Notably, dynamic temporal change in locations of bnl expressing cells was also reported to guide the migrating tracheal progenitors of the larval ASP and the pupal abdominal trachea (Guha et al., 2009; Sato and Kornberg, 2002; Chen and Krasnow, 2014). Dynamic spatiotemporal pattern of bnl-source in these important morphogenetic events can also be created by the migrating population of bnl-expressing cells. In the future, it will be important to investigate how the migration of bnl source may control tracheal growth and how migration of the bnl-source is regulated during development.
Several genetic analyses linked Bnl signaling to embryonic or larval neuronal development (Barrett et al., 2008; Klämbt et al., 1992; Mukherjee et al., 2012). However, in most instances, the identity, morphology, and functional organization of bnl-expressing cells were not well characterized. Employing the genetic strategy combining reporter expression from the bnl-LexA and btl-Gal4 drivers, we identified different types of neurons expressing these genes. In embryos, segmental patterns of bnl expressing neurons were uncovered in the central nervous system. Although Bnl is necessary for larval neuroblast division, the embryonic neuroblast lineage for the bnl-positive neurons was not known. Our analyses of bnl-LexA expression identified the NB 4-4 derived bnl expressing neurons in embryonic CNS. A recent study (Lacin and Truman, 2016) linked the embryonic NB 4-4 lineage to their corresponding post-embryonic progenitors and also showed that the thoracic NB 4-4 lineage generates the leg motor neurons.
In larval eye discs, R8 photoreceptor neurons expressed bnl in a subset of ommatidial clusters. Why do only a subset of retinal ommatidial clusters (Fig. 6A-B) that are located at the dorsal and ventral edges of the discs express bnl is unknown. Although bnl is also expressed in epithelial cells of the eye disc, only the neurons, specifically the R7 photoreceptors that are in close proximity with the bnl-expressing neurons expressed btl. Since btl transcription is a positive feedback response of Bnl signaling, active neuronal exchange of Bnl between the R8 and R7 cells may have induced btl expression in these R7 cells. Both suppression of Btl activity and ligand-independent activation of Btl in the btl-expressing R7 neurons autonomously affected their number and morphology and also resulted in a reciprocal effect on the bnl-expressing R8 photoreceptors (Fig. 6I-M). The abnormal targeting and cell death phenotypes suggested a role of Bnl in maintaining the visual tracks. An earlier study also demonstrated an autonomous defect in the ommatidial clusters in the bnl- and btl- loss of function clones in larval retina (Mukherjee et al., 2012). In contrary to the observation of restricted bnl transcription revealed by various enhancer trap lines or bnl-in situ hybridization (Figs. 6;S4C; Jarecki et al., 1999), the previous report (Mukherjee et al., 2012) hinted at an immunohistochemical detection of low uniform distribution of Bnl protein throughout the larval eye disc. Taken together, these observations may suggest a long range distribution of Bnl from its source.
Finally, our study revealed a fundamental similarity in the intimate organization and interaction of the btl- and bnl- expressing cells in both epithelial and neuronal morphogenesis (Figs. 4,6; Supplementary movies 1,2, 9,10, 12,13). Direct cell-cell contact sites were visualized by the sybGRASP signal between the receptor- and ligand- expressing cells, especially at the growing tip of the embryonic tracheal branches and at the neuronal synapse in the embryonic CNS (Figs. 4L-N; 6S). Since the sybGFP-reconstitution method relies on Ca2+-dependent fusion of synaptobrevin containing vesicles at an active synaptic site (Macpherson et al., 2015), these contacts detected between btl- and bnl- expressing cells may represent the sites of active vesicle exchange. Direct contacts, established by signaling filopodia or cytonemes between the Bnl exchanging cells were previously shown to be essential for signal transduction (Kornberg and Roy, 2014; Roy et al., 2014). Thus, the changing pattern of cell-cell contacts around the growing tracheal branches (Fig 4L-N) or the synaptic contacts in the embryonic midline may represent a spatiotemporal landscape of signaling events between the receptor and the ligand expressing cells. How neurons or epithelial cells exchange Bnl, and how Bnl may guide the cellular patterning of these very different types of cells are currently not known. The new genetic tool and the resource of comprehensive cellular expression and interaction patterns described in this study will greatly facilitate future research to dissect the fundamental mechanisms of Bnl signaling in tissue patterning and remodeling.
Supplementary Material
Figure S1. Specificity and versatility of bnl-LexA expression in all developmental stages. A. RT-PCR analyses on total RNA from bnl-LexA and the original Cas9 flies (control). M, 100 bp Marker (NEB); ∼440 bp RT-PCR amplification band (*) detected from RT-PCR on bnl-lexA RNA. B-I, Genotype: btl-Gal4, UAS-CD8:GFP/+; bnl-LexAlexO-CherryCAAX/+. Bnl expression (red) observed in highly tracheated (green) larval tissues: larval gut (B), ventral muscle (D), central nervous system (E), heart (F); and in adult organs: brain (G), abdominal spiracle (H), muscle/epidermis (I). No expression in larval salivary gland and the organ is not tracheated (C). Scale bars, 100 μm; H, 30 μm.
Figure S2. Expression of bnl-LexA in embryo. A-B′ similar pattern of bnl expression in stage 11 embryo detected by RNA in situ hybridization (A) and by bnl-LexA reporter expression (B, B′). C-C, Stage 13-14 embryos showing similar expression of bnl by RNA in situ hybridization (C) and from bnl-LexA (C). A, C, best focus plane shown; genotype, white. B,B′,C, extended projection of all the optical Z planes; genotype, btl-Gal4, UAS-CD8:GFP/+; bnl-LexA,lexO-CherryCAAX/+. Scale bars, 30 μm (B, B′, C), 50 μm (A, C).
Figure S3. Leg disc associated tracheoblasts cells and bnl expression. A, Bnl expression in leg disc from Fig 5B, shown in red channel. B-C, RNA in situ hybridization marked bnl mRNA in different leg discs. D-D′ bnl-LacZ expression (btl-Gal4, UAS-CD8GFP/+; bnl-lacZ/+) marked nuclei of the bnl-expressing leg disc epithelial cells; bnl expressing cells in disc stalk wrapped the tracheal tube (D); red, immunostaining for anti-pGal. E, bnl-Gal4 expression incompletely marked only some of the bnl-expressing cells in leg disc stalk (bnl-Gal4 X UAS-nlsGFP). red, trachea marked by btlenh-RFPmoe. F, 1151-Gal4 expression in myoblasts (1151-Gal4 X UAS-CD8:GFP) surrounding leg tracheal progenitors (btl>RFPmoe). Myoblasts did not express bnl-LexA. G, 2A12 staining (red) for luminal marking in the leg disc tracheal tube (btl-Gal4>nlsGFP). H, Engrailed immunostaining (green) marked the posterior compartments of the discs in 1st pair of leg discs; leg disc tracheal tubes (btl>RFPmoe) grew along the anterior-posterior compartment border. I, Tracheal connection from TR1 to the 1st pair of leg discs (btl-Gal4 X UAS-nlsGFP). J-K, tracheal overexpression of ligand independent form of activated Btl inhibited the directional growth into the leg discs; red, background auto-fluorescence in 405 nm channel marked tracheal tubes. L, Growth of leg disc tracheal progenitors in mid-pupal leg disc. M, Air sacs and associated leg trachea in the front pair of legs in pharate adult. L-M, btl-Gal4 X UAS-CD8:GFP; arrows, putative adult trachea and air sacs formed by the leg disc tracheal progenitors. N, Schematic drawing of leg trachea and associated ventral air sacs (green) that are likely to be formed from the leg disc tracheal progenitors. E, F, H, J, K; tracheal lumen and discs were imaged with auto fluorescence in 405 nm channel (blue, E, F, H; red; J, K). Scale bars, 30μm. L, M, 100 μm.
Figure S4. Neuronal expression of bnl and btl in larval and embryonic development. A-G, Larval eye disc, anterior - left. A, bnl-Gal4 expression (UAS-CD8:GFP; bnl-Gal4/TM6) in eye disc. B, bnl-lacZ expression (btl-Gal4, UAS-CD8:GFP/+; bnl-lacZ/+) in eye disc. C, bnl RNA in situ hybridization pattern in eye disc showed retinal expression (arrow). D, bnl-expressing cells in eye disc ommatidial neurons (bnl-LexA,lexO-CherryCAAX/+) are marked by Elav antibody staining. E, btl-expressing cells (btl-Gal4, UAS-CD8:GFP/+) in eye disc ommatidia are marked by Elav antibody staining. F, three-dimensional projection showing organization of R8 photoreceptors, marked by Sens immunostaining (blue), bnl- expressing neurons (red), and btl- expressing neurons (green) (btl-Gal4,UAS-CD8:GFP/+; bnl-LexA,lexO-CherryCAAX/+). G, reduction of number of the bnl- (arrows) and btl- (arrowhead) expressing neurons and their axonal projections to the optic lobe when λBtl was expressed in the btl-positive cells (btl-Gal4, UAS-CD8:GFP/UAS-λBtl; bnl-LexA, lexO-CherryCAAX/+). H-M, Stage 14-15 embryo, ventral view, anterior up. H, bnl RNA in situ hybridization pattern in embryonic CNS, arrows, subcellular mRNA localization in the cell body of bnl-expressing neurons. I, midline bnl-expressing cells marked by Elav immunostaining. J, organization of bnl-lacZ marked neuronal nuclei with respect to the btl-expressing midline cells (btl-Gal4, UAS-CD8:GFP/+; bnl-lacZ/+). K-M, bnl-expressing neurons are posterior to the EG neurons (K, eg-Gal4>CD8GFP, bnl-LexA>CherryCAAX) and are not marked by aEn (L) and aHb9 (M) immunostaining. Scale bars, 30 μm.
Supplementary Movies 1-9. Time lapse movies in 3D projections shows dynamic reorganization of bnl-expressing cells (red) and btl (green) expressing trachea during embryonic development (also see Fig 3G). Genotype: btl-Gal4,UAS-CD8:GFP/+; bnl-LexA,lexO-CherryCAAX/+.
Movie 1, middle segments of an embryo from late stage 12-13 onwards.
Movie 2, posterior segments of an embryo from stage 11-12 onwards.
Movie 3, posterior segments of an embryo from stage 11 onwards, only red channel shown marking the bnl-clusters.
Movie 4, tracking of the leading and trailing cells of the bnl-expressing clusters; same embryo as shown in movie #1.
Movie 5, tracking of all the cells in a dorsal cluster, same embryo as shown in movie # 3.
Movie 6, tracking of cells in several lateral clusters in different segments of the embryo shown in movie #2.
Movie 7, migration route shown as an overlay with dots and lines for each cell in a dorsal cluster, embryo same as movie # 3, 5.
Movie 8, migration route shown as an overlay with dots and lines for each cell in lateral cluster, embryo same as movie # 2, 6.
Movie 9, measuring rate of movement by tracking migrating tip of a dorsal branch and the migrating front of the dorsal cluster.
Supplementary Movie 10. Association of the btl expressing tracheoblasts and the bnl expressing cells in 3rd instar leg imaginal disc, views from different angles shown. Genotype: btl-Gal4,UAS-CD8:GFP/+; bnl-LexA,lexO-CherryCAAX/+.
Supplementary Movie 11. Series of Z-stacks of leg tracheoblast cells and associated leg discs (1st pairs) showing EdU incorporation and cell proliferation in ex-vivo cultured organs. Genotype: btl-Gal4,UAS-CD8:GFP/+.
Supplementary Movies 12-13. 3D projection showing organization of bnl- and btl-expressing neurons in eye disc. Genotype: btl-Gal4, UAS-CD8:GFP/+; bnl-LexAlexO-CherryCAAX/+. Blue, anti-Elav staining.
Highlights.
Generation of a bnl-LexA/lexO based targeted expression system by genome editing
bnl-LexA expressed target-specifically and identified new bnl expressing cells
Mutant analyses validated the functional role of newly identified bnl-expressions
Tracking bnl-LexA positive live embryonic cells identified their dynamic migration
Embryonic bnl-sources migrate in synchrony with tracheal branch and dorsal closure
Acknowledgments
We thank Dr. T.B. Kornberg, Dr. L. Jiang, Dr. H. Bellen, Dr. H. Lacin, Dr. J. Skeath, Dr. L. Pick, Dr. I. Ando, Dr. W.F. Odenwald, the Bloomington Stock Center, the Developmental Studies Hybridoma Bank, and the PACMAN resource center for reagents; Dr. A.E. Beaven and the UMD Imaging core facility for confocal imaging; Dr. H. Lacin and Dr. J. Skeath for discussions and reagents; Dr. L. Pick and Dr. N. Andrews for comments on the manuscript. Funding from: NIH R00HL114867 and UMD Research and Scholarship Award (RASA) to S.R.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmad SM, Baker BS. Sex-specific deployment of FGF signaling in Drosophila recruits mesodermal cells into the male genital imaginal disc. Cell. 2002;109:651–661. doi: 10.1016/s0092-8674(02)00744-4. [DOI] [PubMed] [Google Scholar]
- Bae YK, Trisnadi N, Kadam S, Stathopoulos A. The role of FGF signaling in guiding coordinate movement of cell groups. Cell Adhesion & Migration. 2014;6:397–403. doi: 10.4161/cam.21103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett AL, Krueger S, Datta S. Branchless and Hedgehog operate in a positive feedback loop to regulate the initiation of neuroblast division in the Drosophila larval brain. Dev Biol. 2008;317:234–245. doi: 10.1016/j.ydbio.2008.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beenken A, Mohammadi M. The FGF family: biology, pathophysiology and therapy. Nat Rev Drug Discov. 2009;8:235–253. doi: 10.1038/nrd2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer KJ, Trautman JK, Mukherjee K, Carroll D. Donor DNA Utilization during Gene Targeting with Zinc-finger Nucleases. G3 (Bethesda) 2013;3:657–664. doi: 10.1534/g3.112.005439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161:1169–1175. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenstein D. The Postembryonic Development of Biology of Drosophila. In: Demerec M, editor. Hafner, Drosophila. New York: 1965. pp. 275–367. [Google Scholar]
- Brewer JR, Mazot P, Soriano P. Genetic insights into the mechanisms of Fgf signaling. Genes & Development. 2016;30:751–771. doi: 10.1101/gad.277137.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadus J, Skeath JB, Spana EP, Bossing T, Technau G, Doe CQ. New neuroblast markers and the origin of the aCC/pCC neurons in the Drosophila central nervous system. Mechanisms of Development. 1995;53:393–402. doi: 10.1016/0925-4773(95)00454-8. [DOI] [PubMed] [Google Scholar]
- Centanin L, Dekanty A, Romero N, Irisarri M, Gorr TA, Wappner P. Cell Autonomy of HIF Effects in Drosophila: Tracheal Cells Sense Hypoxia and Induce Terminal Branch Sprouting. Developmental Cell. 2008;14:547–558. doi: 10.1016/j.devcel.2008.01.020. [DOI] [PubMed] [Google Scholar]
- Chen F, Krasnow MA. Progenitor outgrowth from the niche in Drosophila trachea is guided by FGF from decaying branches. Science. 2014;343:186–189. doi: 10.1126/science.1241442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CQ. Molecular markers for identified neuroblasts and ganglion mother cells in the Drosophila central nervous system. Development. 1992;116:855–863. doi: 10.1242/dev.116.4.855. [DOI] [PubMed] [Google Scholar]
- Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, Sullender M, Ebert BL, Xavier RJ, Root DE. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol. 2014;32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohl DM, Silies MA, Gao XJ, Bhalerao S, Luongo FJ, Lin CC, Potter CJ, Clandinin TR. A versatile in vivo system for directed dissection of gene expression patterns. Nat Meth. 2011;8:231–237. doi: 10.1038/nmeth.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González M, Martín-Ruíz I, Jiménez S, Pirone L, Barrio R, Sutherland JD. Generation of stable Drosophila cell lines using multicistronic vectors. Sci Rep. 2011;1:1–7. doi: 10.1038/srep00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, O'Connor-Giles KM. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics. 2014;196:961–971. doi: 10.1534/genetics.113.160713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha A, Lin L, Kornberg TB. Regulation of Drosophila matrix metalloprotease Mmp2 is essential for wing imaginal disc:trachea association and air sac tubulogenesis. Dev Biol. 2009;335:317–326. doi: 10.1016/j.ydbio.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haack T, Schneider M, Schwendele B, Renault AD. Drosophila heart cell movement to the midline occurs through both cell autonomous migration and dorsal closure. Dev Biol. 2014;396:169–182. doi: 10.1016/j.ydbio.2014.08.033. [DOI] [PubMed] [Google Scholar]
- Higashijima S, Shishido E, Matsuzaki M, Saigo K. eagle, a member of the steroid receptor gene superfamily, is expressed in a subset of neuroblasts and regulates the fate of their putative progeny in the Drosophila CNS. Development. 1996;122:527–536. doi: 10.1242/dev.122.2.527. [DOI] [PubMed] [Google Scholar]
- Jarecki J, Johnson E, Krasnow MA. Oxygen regulation of airway branching in Drosophila is mediated by branchless FGF. Cell. 1999;99:211–220. doi: 10.1016/s0092-8674(00)81652-9. [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klämbt C, Glazer L, Shilo BZ. breathless, a Drosophila FGF receptor homolog, is essential for migration of tracheal and specific midline glial cells. Genes & Development. 1992;6:1668–1678. doi: 10.1101/gad.6.9.1668. [DOI] [PubMed] [Google Scholar]
- Kornberg TB, Roy S. Communicating by touch--neurons are not alone. Trends Cell Biol. 2014;24:370–376. doi: 10.1016/j.tcb.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacin H, Truman JW. Lineage mapping identifies molecular and architectural similarities between the larval and adult Drosophila central nervous system. Elife. 2016;5:e13399. doi: 10.7554/eLife.13399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai SL, Lee T. Genetic mosaic with dual binary transcriptional systems in Drosophila. Nat Neurosci. 2006;9:703–709. doi: 10.1038/nn1681. [DOI] [PubMed] [Google Scholar]
- Li W, Köster J, Xu H, Chen CH, Xiao T, Liu JS, Brown M, Liu XS. Quality control, modeling, and visualization of CRISPR screens with MAGeCK-VISPR. Genome Biol. 2015;16:281. doi: 10.1186/s13059-015-0843-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CC, Potter CJ. Editing Transgenic DNA Components by Inducible Gene Replacement in Drosophila melanogaster. Genetics. 2016;203:1613–1628. doi: 10.1534/genetics.116.191783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson LJ, Zaharieva EE, Kearney PJ, Alpert MH, Lin TY, Turan Z, Lee CH, Gallio M. Dynamic labelling of neural connections in multiple colours by trans-synaptic fluorescence complementation. Nat Comms. 2015;6:10024. doi: 10.1038/ncomms10024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning G, Krasnow MA. Development of the Drosophila tracheal system. In: Bate M, Arias AM, editors. The Development of Drosophila melanogaster. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 1993. pp. 609–685. [Google Scholar]
- Mukherjee T, Choi I, Banerjee U. Genetic analysis of fibroblast growth factor signaling in the Drosophila eye. G3 (Bethesda) 2012;2:23–28. doi: 10.1534/g3.111.001495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Ngo TTB, Hibbard KL, Murphy C, Jenett A, Truman JW, Rubin GM. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186:735–755. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port F, Chen HM, Lee T, Bullock SL. An optimized CRISPR/Cas toolbox for efficient germline and somatic genome engineering in Drosophila. 2014 doi: 10.1101/003541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Huang H, Liu S, Kornberg TB. Cytoneme-mediated contact-dependent transport of the Drosophila decapentaplegic signaling protein. Science. 2014;343:1244624–1244624. doi: 10.1126/science.1244624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Kornberg TB. Paracrine signaling mediated at cell-cell contacts. Bioessays. 2015;37:25–33. doi: 10.1002/bies.201400122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Kornberg TB. FGF is an essential mitogen and chemoattractant for the air sacs of the drosophila tracheal system. Developmental Cell. 2002;3:195–207. doi: 10.1016/s1534-5807(02)00202-2. [DOI] [PubMed] [Google Scholar]
- Stathopoulos A, Tam B, Ronshaugen M, Frasch M, Levine M. pyramus and thisbe: FGF genes that pattern the mesoderm of Drosophila embryos. Genes & Development. 2004;18:687–699. doi: 10.1101/gad.1166404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland D, Samakovlis C, Krasnow MA. branchless encodes a Drosophila FGF homolog that controls tracheal cell migration and the pattern of branching. Cell. 1996;87:1091–1101. doi: 10.1016/s0092-8674(00)81803-6. [DOI] [PubMed] [Google Scholar]
- Triantafyllou A, Liakos P, Tsakalof A, Georgatsou E, Simos G, Bonanou S. Cobalt induces hypoxia-inducible factor-1 alpha (HIF-1 alpha) in HeLa cells by an iron-independent, but ROS-, PI-3K- and MAPK-dependent mechanism. Free Radic Res. 2006;40:847–856. doi: 10.1080/10715760600730810. [DOI] [PubMed] [Google Scholar]
- Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Vol. 10. Nature Publishing Group; 2010. pp. 116–129. [DOI] [PubMed] [Google Scholar]
- Wu D, Yotnda P. Induction and testing of hypoxia in cell culture. J Vis Exp. 2011 doi: 10.3791/2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Gustafson WC, Han C, Lafaye C, Noirclerc-Savoye M, Ge WP, Thayer DA, Huang H, Kornberg TB, Royant A, Jan LY, Jan YN, Weiss WA, Shu X. An improved monomeric infrared fluorescent protein for neuronal and tumour brain imaging. Nat Comms. 2014;5:3626. doi: 10.1038/ncomms4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zartman J, Restrepo S, Basler K. A high-throughput template for optimizing Drosophila organ culture with response-surface methods. Development. 2013;140:667–674. doi: 10.1242/dev.088872. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Specificity and versatility of bnl-LexA expression in all developmental stages. A. RT-PCR analyses on total RNA from bnl-LexA and the original Cas9 flies (control). M, 100 bp Marker (NEB); ∼440 bp RT-PCR amplification band (*) detected from RT-PCR on bnl-lexA RNA. B-I, Genotype: btl-Gal4, UAS-CD8:GFP/+; bnl-LexAlexO-CherryCAAX/+. Bnl expression (red) observed in highly tracheated (green) larval tissues: larval gut (B), ventral muscle (D), central nervous system (E), heart (F); and in adult organs: brain (G), abdominal spiracle (H), muscle/epidermis (I). No expression in larval salivary gland and the organ is not tracheated (C). Scale bars, 100 μm; H, 30 μm.
Figure S2. Expression of bnl-LexA in embryo. A-B′ similar pattern of bnl expression in stage 11 embryo detected by RNA in situ hybridization (A) and by bnl-LexA reporter expression (B, B′). C-C, Stage 13-14 embryos showing similar expression of bnl by RNA in situ hybridization (C) and from bnl-LexA (C). A, C, best focus plane shown; genotype, white. B,B′,C, extended projection of all the optical Z planes; genotype, btl-Gal4, UAS-CD8:GFP/+; bnl-LexA,lexO-CherryCAAX/+. Scale bars, 30 μm (B, B′, C), 50 μm (A, C).
Figure S3. Leg disc associated tracheoblasts cells and bnl expression. A, Bnl expression in leg disc from Fig 5B, shown in red channel. B-C, RNA in situ hybridization marked bnl mRNA in different leg discs. D-D′ bnl-LacZ expression (btl-Gal4, UAS-CD8GFP/+; bnl-lacZ/+) marked nuclei of the bnl-expressing leg disc epithelial cells; bnl expressing cells in disc stalk wrapped the tracheal tube (D); red, immunostaining for anti-pGal. E, bnl-Gal4 expression incompletely marked only some of the bnl-expressing cells in leg disc stalk (bnl-Gal4 X UAS-nlsGFP). red, trachea marked by btlenh-RFPmoe. F, 1151-Gal4 expression in myoblasts (1151-Gal4 X UAS-CD8:GFP) surrounding leg tracheal progenitors (btl>RFPmoe). Myoblasts did not express bnl-LexA. G, 2A12 staining (red) for luminal marking in the leg disc tracheal tube (btl-Gal4>nlsGFP). H, Engrailed immunostaining (green) marked the posterior compartments of the discs in 1st pair of leg discs; leg disc tracheal tubes (btl>RFPmoe) grew along the anterior-posterior compartment border. I, Tracheal connection from TR1 to the 1st pair of leg discs (btl-Gal4 X UAS-nlsGFP). J-K, tracheal overexpression of ligand independent form of activated Btl inhibited the directional growth into the leg discs; red, background auto-fluorescence in 405 nm channel marked tracheal tubes. L, Growth of leg disc tracheal progenitors in mid-pupal leg disc. M, Air sacs and associated leg trachea in the front pair of legs in pharate adult. L-M, btl-Gal4 X UAS-CD8:GFP; arrows, putative adult trachea and air sacs formed by the leg disc tracheal progenitors. N, Schematic drawing of leg trachea and associated ventral air sacs (green) that are likely to be formed from the leg disc tracheal progenitors. E, F, H, J, K; tracheal lumen and discs were imaged with auto fluorescence in 405 nm channel (blue, E, F, H; red; J, K). Scale bars, 30μm. L, M, 100 μm.
Figure S4. Neuronal expression of bnl and btl in larval and embryonic development. A-G, Larval eye disc, anterior - left. A, bnl-Gal4 expression (UAS-CD8:GFP; bnl-Gal4/TM6) in eye disc. B, bnl-lacZ expression (btl-Gal4, UAS-CD8:GFP/+; bnl-lacZ/+) in eye disc. C, bnl RNA in situ hybridization pattern in eye disc showed retinal expression (arrow). D, bnl-expressing cells in eye disc ommatidial neurons (bnl-LexA,lexO-CherryCAAX/+) are marked by Elav antibody staining. E, btl-expressing cells (btl-Gal4, UAS-CD8:GFP/+) in eye disc ommatidia are marked by Elav antibody staining. F, three-dimensional projection showing organization of R8 photoreceptors, marked by Sens immunostaining (blue), bnl- expressing neurons (red), and btl- expressing neurons (green) (btl-Gal4,UAS-CD8:GFP/+; bnl-LexA,lexO-CherryCAAX/+). G, reduction of number of the bnl- (arrows) and btl- (arrowhead) expressing neurons and their axonal projections to the optic lobe when λBtl was expressed in the btl-positive cells (btl-Gal4, UAS-CD8:GFP/UAS-λBtl; bnl-LexA, lexO-CherryCAAX/+). H-M, Stage 14-15 embryo, ventral view, anterior up. H, bnl RNA in situ hybridization pattern in embryonic CNS, arrows, subcellular mRNA localization in the cell body of bnl-expressing neurons. I, midline bnl-expressing cells marked by Elav immunostaining. J, organization of bnl-lacZ marked neuronal nuclei with respect to the btl-expressing midline cells (btl-Gal4, UAS-CD8:GFP/+; bnl-lacZ/+). K-M, bnl-expressing neurons are posterior to the EG neurons (K, eg-Gal4>CD8GFP, bnl-LexA>CherryCAAX) and are not marked by aEn (L) and aHb9 (M) immunostaining. Scale bars, 30 μm.
Supplementary Movies 1-9. Time lapse movies in 3D projections shows dynamic reorganization of bnl-expressing cells (red) and btl (green) expressing trachea during embryonic development (also see Fig 3G). Genotype: btl-Gal4,UAS-CD8:GFP/+; bnl-LexA,lexO-CherryCAAX/+.
Movie 1, middle segments of an embryo from late stage 12-13 onwards.
Movie 2, posterior segments of an embryo from stage 11-12 onwards.
Movie 3, posterior segments of an embryo from stage 11 onwards, only red channel shown marking the bnl-clusters.
Movie 4, tracking of the leading and trailing cells of the bnl-expressing clusters; same embryo as shown in movie #1.
Movie 5, tracking of all the cells in a dorsal cluster, same embryo as shown in movie # 3.
Movie 6, tracking of cells in several lateral clusters in different segments of the embryo shown in movie #2.
Movie 7, migration route shown as an overlay with dots and lines for each cell in a dorsal cluster, embryo same as movie # 3, 5.
Movie 8, migration route shown as an overlay with dots and lines for each cell in lateral cluster, embryo same as movie # 2, 6.
Movie 9, measuring rate of movement by tracking migrating tip of a dorsal branch and the migrating front of the dorsal cluster.
Supplementary Movie 10. Association of the btl expressing tracheoblasts and the bnl expressing cells in 3rd instar leg imaginal disc, views from different angles shown. Genotype: btl-Gal4,UAS-CD8:GFP/+; bnl-LexA,lexO-CherryCAAX/+.
Supplementary Movie 11. Series of Z-stacks of leg tracheoblast cells and associated leg discs (1st pairs) showing EdU incorporation and cell proliferation in ex-vivo cultured organs. Genotype: btl-Gal4,UAS-CD8:GFP/+.
Supplementary Movies 12-13. 3D projection showing organization of bnl- and btl-expressing neurons in eye disc. Genotype: btl-Gal4, UAS-CD8:GFP/+; bnl-LexAlexO-CherryCAAX/+. Blue, anti-Elav staining.


