Figure 2. Is there a ribosome code?
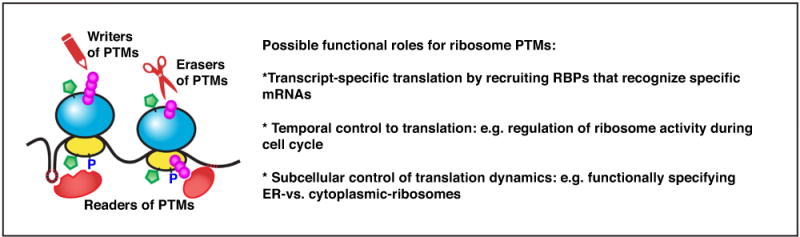
Diverse PTMs observed in RPs include O-GlcNAcylation (represented by green pentagons), phosphorylation, and ubiquitylation. The enzymes that can catalyze (writers) or reverse (erasers) the PTMs may interact with and modify different sub-pools of ribosomes, leading to increased dynamics of ribosome heterogeneity. Analogous to the ‘histone code’, the possibility of downstream trans-factors that can recognize combinations of these PTMs (readers) remains to be investigated. It is possible to envision that PTMs can recruit RNA binding proteins (RBPs) to the ribosome that can specifically recognize certain structures or sequence motifs within selective groups of mRNAs.
