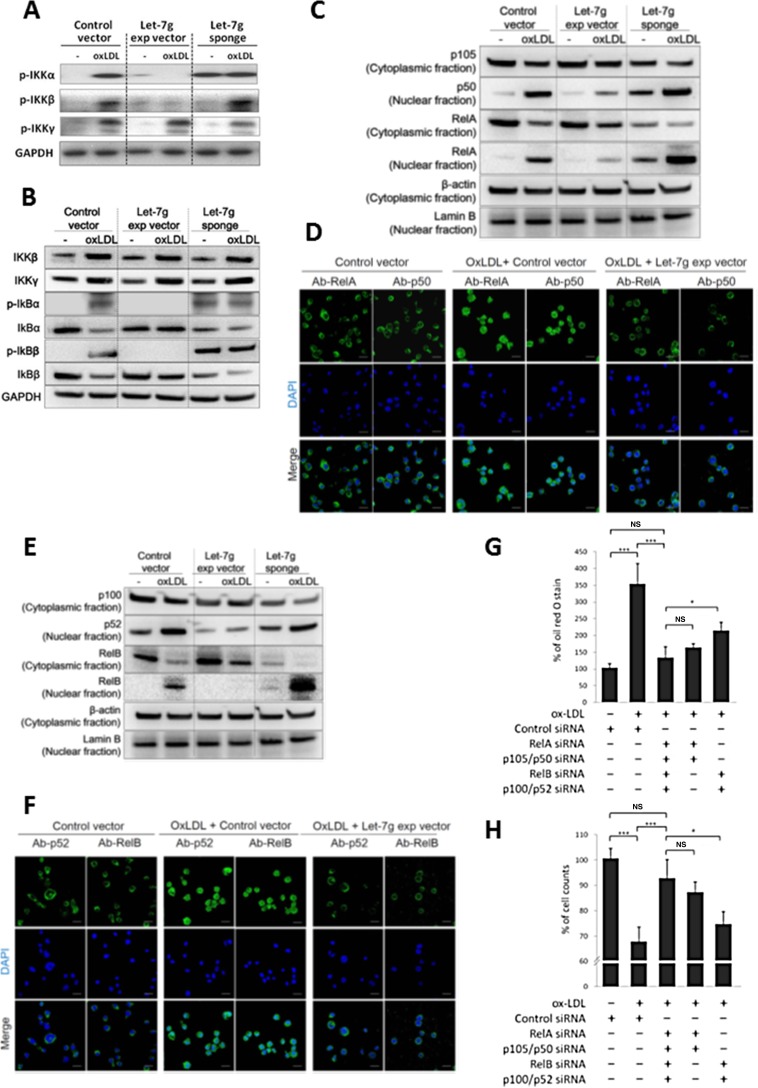
Figure 4. Let-7g inhibits canonical and non-canonical NF-κB signaling pathways.
Macrophages were differentiated from THP-1 cells and were infected with LV-let7g, LV-let7g-sponge or control lentivirus. (A) Western blot analysis for phosphorylated- IKKα, phosphorylated-IKKβ and phosphorylated-IKKγ. GAPDH was served as a loading control. (B) Western blot analysis for IKKβ, IKKγ, IκBα, IκBβ, phosphorylated-IκBα (p-IκBα) and p-IκBβ. GAPDH was served as a loading control. (C) Cellular distribution of transcription factors in the canonical pathway. p105, p50 and RelA in the cytoplasm and nucleus with cytoplasmic β-actin and nuclear lamin B as the respective markers. (D) The confocal laser-scanning microscopy revealed the localization of fluorescently tagged p50 and RelA. Coloring indicated GFP (green) and the DNA dye DAPI (blue). Scale bar: 20 μm. (E) Cellular distribution of transcription factors in the non-canonical pathway. p100, p52 and RelB proteins in the cytoplasm and nucleus with cytoplasmic β-actin and nuclear lamin B as the respective markers. (F) The confocal laser-scanning microscopy showed the localization of GFP-tagged p52 and RelB proteins. Scale bar: 20 μm. (G, H). Knockdown of the canonical components (RelA and p105, 5th bar), or non-canonical components (RelB and p52, 4th bar) caused different effects on (G) intracellular lipid accumulation and (H) cell counts in macrophages. The data were from at least three independent experiments. Data in each bar chart are presented as mean ± SEM *P < 0.05, **P < 0.01, ***P < 0.001.