Figure 1.
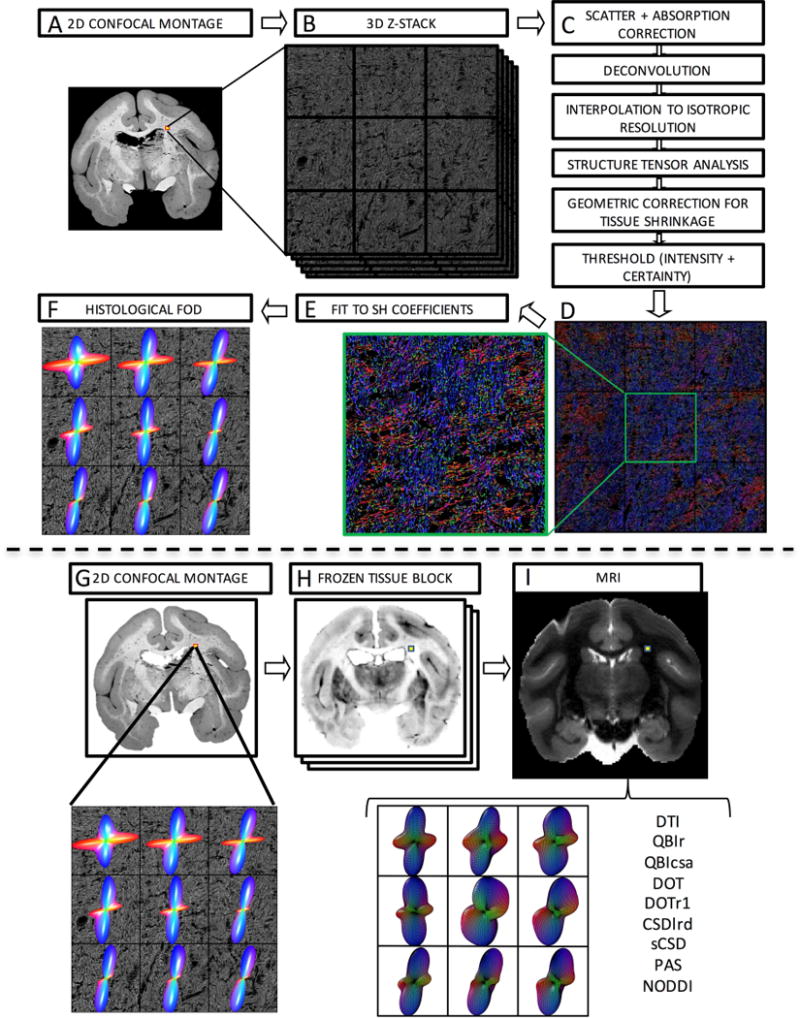
Histological procedures (top) and image registration (bottom). Confocal acquisition includes a 2D low-resolution montage (A) and a high-resolution 3D z-stack (B). Image pre-processing (C) comprises light scatter and absorption correction, deconvolution, and interpolation, followed by structure tensor analysis. This is followed by geometric correction for tissue shrinkage and thresholding fibers. This results in an orientation estimate for every pixel in the z-stack occupied by a fiber, here shown as an RGB color map (D) where red, green, and blue represent fiber oriented right/left, anterior/posterior, and superior/inferior. Zooming in on the center voxel shows crossing fibers oriented primarily left/right and superior/inferior. Fitting the orientation distribution to spherical harmonic coefficients (E) results in the histologically defined ground truth FODs, displayed as 3D glyphs (F). The registration procedure involves 2D registration of the 2D confocal montage (G) to the corresponding frozen tissue block (H) and subsequent 3D registration to the non-diffusion weighted image (I). From this, the signal corresponding to the high-resolution z-stack can be determined and processed using a chosen reconstruction method for direct voxel-wise comparison of histology and dMRI.
