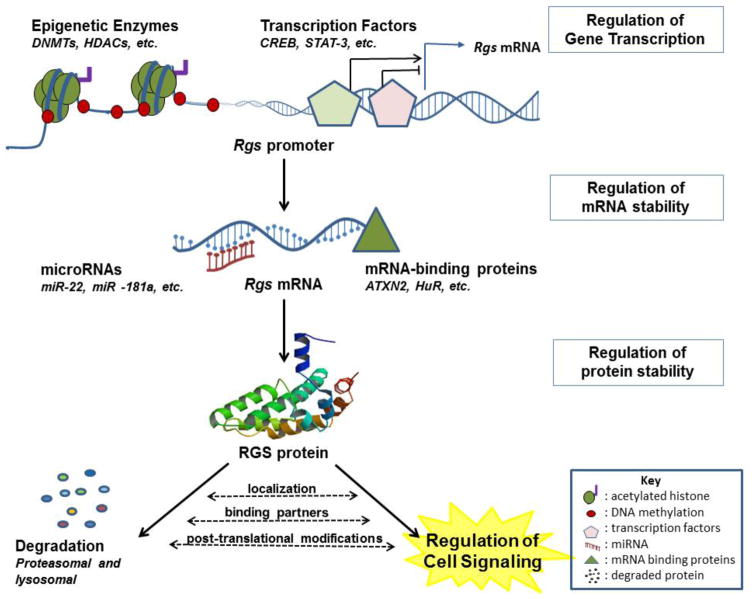
Figure 1. RGS gene expression and protein stability are regulated by multiple mechanisms.
Multiple regulatory mechanisms participate in determining the level of RGS proteins. Epigenetic modifications, mainly histone deacetylation and DNA methylation that are mediated by histone deacetylases (HDACs) or DNA methyltransferases (DNMTs), tighten the chromatin structure at RGS genes promoters, thereby obstructing the access of transcription factors and other proteins that are essential for transcription initiation, ultimately resulting in suppression of RGS gene expression. Multiple transcription factors directly bind RGS genes promoters to activate or repress transcription, adding another layer of regulation to the expression of RGS genes. In addition to transcription regulation, RGS mRNAs are targeted by both microRNAs and mRNA-binding proteins to either degrade or stabilize the respective mRNA, which critically determines the final levels of translated RGS proteins. Finally, active RGS proteins participate in various G protein dependant and independent signalling pathways in different cellular compartments. The activity and stability of RGS proteins is influenced by post-translational modifications such as phosphorylation, association with specific binding partners, and cellular localization of RGS proteins. Many RGS proteins undergo proteasomal degradation while some are degraded via lysosomal degradation. Regardless, this degradation is a critical step of regulation that ultimately governs the level of active cellular RGS proteins at a given time.