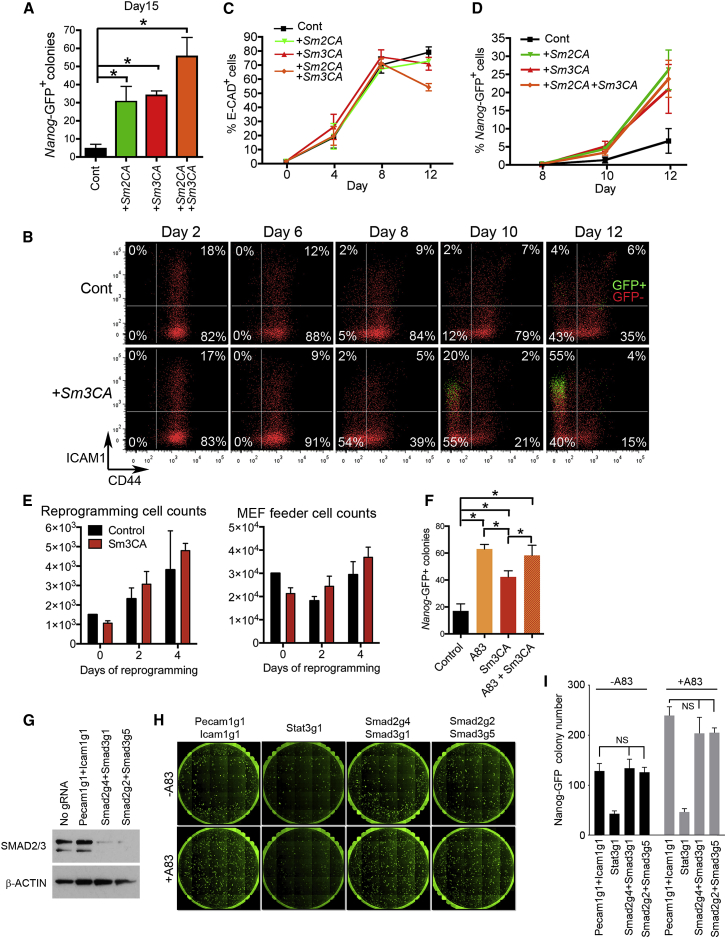
Figure 3.
Constitutively Active Smad2/3 Boosts Reprogramming
(A) Nanog-GFP+ colony numbers on day 15 of reprogramming with retroviral infection of control vector (Cont), constitutive active Smad2 (+Sm2CA), Smad3 (+Sm3CA), or Smad2CA plus Smad3CA (+Sm2CA+Sm3CA).
(B) CD44/ICAM1/Nanog-GFP expression during reprogramming with control (top) and Smad3CA (bottom) expression vector infection. Red, Nanog-GFP−; green, Nanog-GFP+ Tg cells.
(C and D) E-CAD (C) and Nanog-GFP (D) expression during reprogramming with control, Smad2CA, Smad3CA, or Smad2CA plus Smad3CA expression vector infection.
(E) Numbers of Tg (left) and wild-type feeder MEFs (right) with control (Cont) or Sm3CA vectors during reprogramming.
(F) Nanog-GFP+ colony numbers on day 15 of reprogramming in the presence of A83, Sm3CA, or A83 plus Sm3CA.
(G) Efficient double knockout (dKO) of Smad2 and Smad3 was observed 72 hr after co-infection of lentiviruses encoding gRNA targeting Smad2 and Smad3 (Smad2g2+Smad3g1, Smad2g4+Smad3g5). Controls included samples with no gRNA infection and with co-infection of gRNA viruses against Pecam1 (Pecam1g1) and Icam1 (Icam1g1), respectively.
(H and I) Smad2/3 double KO did not affect reprogramming efficiency in the presence or absence of A83. Pecam1g1+Icam1g1 and Stat3g1 (gRNA against Stat3) were used as negative and positive controls, respectively.
Graphs represent averages of 3 (A, C, D–F, F) or 2 (I) independent experiments with 2 technical replicates. Error bars indicate SD. ∗p < 0.05; NS, not significant (two-sided t test). See also Figure S3.