Fig. 1.
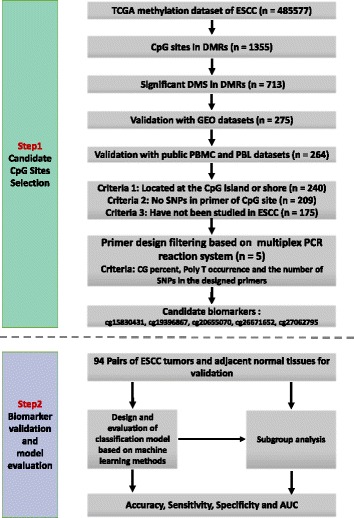
Flow chart of the study design. Candidate biomarkers were selected from the high-throughput DNA methylation microarrays from the TCGA project and further validated with the ESCC methylation data from the GEO dataset, as well as PBMC and PBL from healthy controls. In addition, the PBL and PBMC methylation datasets from healthy samples were also utilized for biomarker filtering. Based on our preliminary screening, the candidate methylation biomarkers for ESCC were then further validated with targeted bisulfite sequencing in independent Chinese Han ESCC patients
