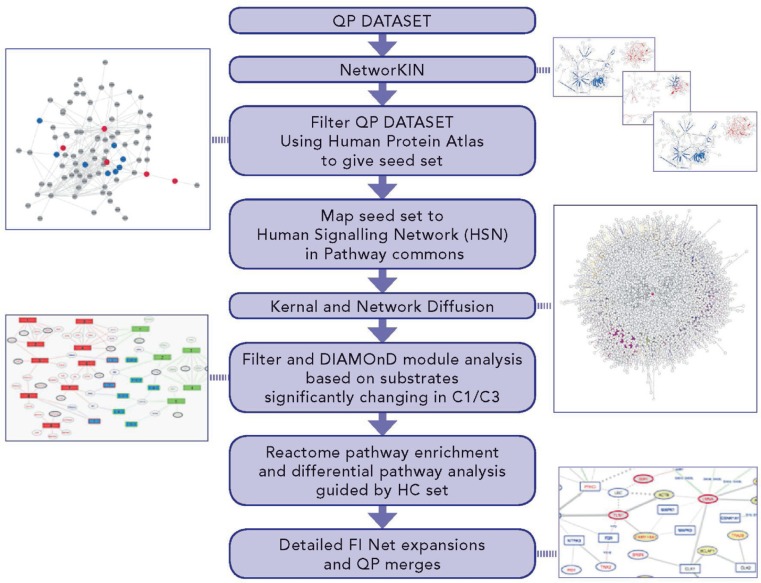
Figure 4. Differential pathway and network analysis process - DRAGON protocol - to identify networks specific to either FUS or WT signaling.
DRAGON uniquely expands and filters protein interaction networks. Depicted multiple steps cover: (i) expansion of observed altered phosphosites to include predicted kinases; (ii) filtering by tissue expression; (iii) seeding and expansion of a general PPI network (the HSN) through a graph-kernel to create an effective network focused on each comparison; (iv) expansion of the proteins in each comparison using a module detection algorithm to give sets of proteins significantly implicated in signalling events; (v) enrichment in module sets for Reactome pathways; (vi) filtering by subtraction of pathway lists to remove identical hits in each comparison; (vii) filtering differential pathways to exclude those without at least one HC protein and (viii) detailed functional network expansion around specific pathways.