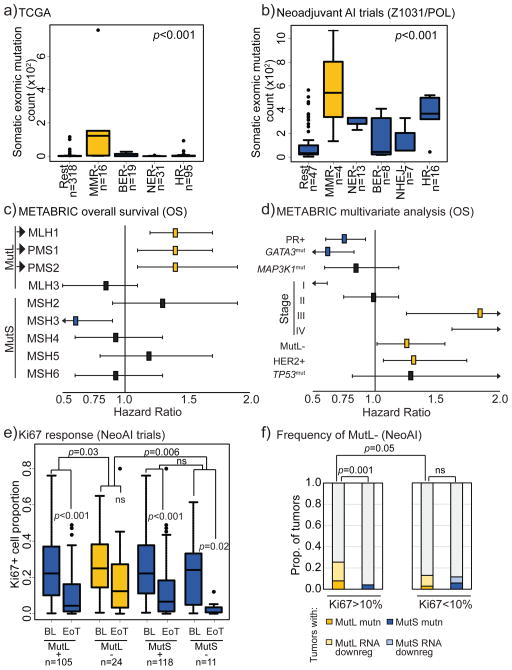
Figure 1. Mismatch repair dysregulation associates with high mutation load and poor clinical outcome in patients with ER+ breast cancer.
a+b) ER+ tumors with non-synonymous mutations in pathway-unique MMR genes from TCGA (a) and NeoAI (b) datasets have increased overall exomic mutation load compared to tumors without. BER, base excision repair; NER, nucleotide excision repair; HR, homologous recombination; NHEJ, non-homologous end joining; Rest, all others. ANOVA with Tukey comparison determined p-values. c+d) Forest plots demonstrating that ER+ tumors with low MutL gene mRNA (<mean-1.5xStDev) in METABRIC associate selectively with poor overall survival (c) and that low MutL mRNA is an independent prognosticator of poor survival in ER+ breast cancer (d). Log rank test was used to determine significance and Cox Regression Proportional Hazards for multivariate analysis. Benjamini-Hochberg correction was applied for multiple comparisons. e) Box plots demonstrating that ER+ tumors from NeoAI (Z1031, POL, NeoPalAna) with mutations in and/or low expression of MutL genes are significantly more likely to be resistant to endocrine therapy as determined by change in Ki67 levels on AI treatment. Friedman Rank Sum test for repeated measures determined p-values for pair-wise comparisons, and Kruskal Wallis test determined p-value for between-group comparisons. f) Stacked column graph depicting frequency of incidence of MutL dysregulation in ER+ tumors from NeoAI. Fisher’s Exact test determined p-values. Accompanying information presented in Figure S1 and Table 1. Horizontal lines indicate mean and error bars denote standard error for box plots.