Abstract
Cilia are essential to many diverse cellular processes. Although many major axonemal components have been identified and studied, how they interact to form a functional axoneme is not completely understood. To further our understanding of the protein composition of human airway cilia, we performed a semi-quantitative analysis of ciliary axonemes using label-free LC/MSE which identified over 400 proteins with high confidence. Tubulins were the most abundant proteins identified, with evidence for 20 different isoforms obtained. Twelve different isoforms of axonemal dynein heavy chain were also identified. Absolute quantification of the non-tubulin components demonstrated a greater than 75-fold range of protein abundance (RSPH9;1850 fmol vs CCDC103;24 fmol), adding another level of complexity to axonemal structure. Of the identified proteins, approximately 70% are known axonemal proteins. In addition, many previously uncharacterized proteins were identified. Unexpectedly, several of these, including ERICH3, C1orf87, and CCDC181, were present at high relative abundance in the cilia. RT-PCR analysis and immunoblotting confirmed cilia specific expression for eight uncharacterized proteins, and fluorescent microscopy demonstrated unique axonemal localizations. These studies have provided the first quantitative analysis of the ciliary proteome and have identified and characterized several previously unknown proteins as major constituents of human airway cilia.
Keywords: cilia, axoneme, mass spectrometry, proteomics, airway, dynein, tubulin, radial spoke
Graphical abstract
Introduction
The cilia that line the human respiratory tract provide the force necessary for efficient mucociliary clearance. By their continuous and coordinated beating, cilia transport mucus and entrapped pathogens and toxins out of the airways, helping to maintain the lung in a healthy state (1–3). Impaired mucociliary clearance is a feature of many airway diseases, including cystic fibrosis (CF) and primary ciliary dyskinesia (PCD). Cystic fibrosis is caused by mutations in the CFTR gene (4, 5), which encodes an ATP regulated anion channel that helps to maintain the proper hydration of the airway surface. In the absence of CFTR function, the mucus layer becomes dehydrated, ciliary movement is impaired, and mucociliary clearance fails (6). PCD is a genetically heterogeneous disease, with causative mutations in over 30 genes identified (7). Many of these mutations are in genes that encode structural proteins of the axoneme itself (e.g., DNAI1, DNAH5) (8, 9) but others encode cytoplasmic assembly or regulatory proteins (e.g., SPAG1, MCIDAS, HEATR2 (10–12). These mutations impair ciliary function and result in impaired or absent mucociliary clearance. Both of these diseases result in frequent and persistent episodes of lung infection, eventually causing bronchiectasis and lung failure (6, 7). Thus studies of mucociliary clearance and the structure and function of cilia have the potential to improve the clinical outcome of patients with these and other debilitating lung diseases.
The cilia themselves are composed of a highly conserved microtubule structure, the axoneme, consisting of 9 microtubule doublets arranged in a circular pattern around a central pair of single microtubules (the 9+2 axoneme). This structure is conserved among most motile cilia and flagella, while the core axonemal structure of primary cilia and the cilia at the embryonic node consists of 9 outer doublets without a central pair (9+0 axoneme). The basic axonemal structure is formed primarily of alpha and beta tubulin, but each complete cilium is composed of several hundred additional proteins that are required for the efficient functioning of this organelle. The composition and structure of the many different specialized axonemes differ between both species and tissues. For example, human airway and most other motile cilia possess three radial spoke head structures, while the well-studied Chlamydomonas flagellum lacks the third radial spoke head (13, 14). Further, the flagella found on sperm (15, 16), the cilia of retinal photoreceptor cells (17), and the cilia present in the airway (18), each contain a unique combination of proteins required to fulfill their specific function. It has also been recognized that the structure of the ciliary axoneme is not uniform along its length, and so the composition and abundance of ciliary proteins varies depending on the location within the cilium. This further adds to the complexity of this organelle. For example, in motile cilia of the airway, the outer dynein arm heavy chain DNAH5 is found along the entire length of the cilium, while DNAH11 is found only proximally and DNAH9 is found only in the distal portion of the cilium (19, 20).
To fully understand the molecular mechanisms that control ciliary beat frequency, waveform, and coordination, it will be necessary to first identify the complete ciliary proteome. Several investigators have employed both genetic and proteomic techniques to begin to identify the entire complement of ciliary proteins. For example, proteomic approaches have been used to analyze primary cilia from human retinal pigmented epithelial cells, mouse kidney, fibroblast, and photoreceptor cells, rat olfactory cells, swine choroid plexus epithelial cells (17, 21–24), and flagella from Chlamydomonas (25). Comparison of gene expression patterns from tissues with or without cilia was utilized to identify cilia specific genes in mouse (26), and a comparative genomics approach was used to identify over 600 genes present in organisms with cilia or flagella (27). Whole genome expression sequencing of bronchial biopsies identified over 200 potential ciliary genes (28). Specifically in airway epithelial cells, we previously performed a liquid chromatography-tandem mass spectrometry (LC/MS/MS)-based proteomic analysis of axonemes isolated from human bronchial epithelial cells to identify over 200 known and potential ciliary proteins (18). While these and other studies have contributed greatly to our knowledge of ciliary composition, no single technique is capable of providing a complete understanding of the ciliary proteome. While RNA expression or genetic approaches can identify hundreds of genes expressed in a “cilia-specific” pattern, they provide no evidence that these proteins are actually made, or whether they are part of the ciliary axoneme itself or play a role in the assembly or function of the axoneme. Proteomic studies are specifically designed to identify proteins that are present in the isolated organelle, but these are limited by the purity of the preparation and the sensitivity of the method. Further, none of the above approaches have yet provided any quantitative data on the relative abundance of the individual protein components.
Recent advances in mass spectrometry (MS) hardware and data acquisition strategies have enabled more sensitive and comprehensive analysis of complex proteomic samples. One of these strategies known generically as data-independent acquisition (DIA) circumvents the stochastic sampling and resulting intensity biases inherent in data-dependent acquisition (DDA) approaches by fragmenting all precursor ions eluting from an LC separation without regard for intensity, enabling effective interrogation of lower abundance species (29). Unlike DDA approaches which are serial in nature and generally target the most intense signals in a given precursor spectrum, DIA fragments all precursors present in the collision cell in parallel, independent of intensity or other individual precursor attributes. A number of variations of DIA approaches exist, some of which are unique to specific hardware vendors, DIA approaches such as PAcIFIC (30) or SWATH-MS (31) utilize sets of relatively large (tens of Th), overlapping mass ranges where precursors are cofragmented in the collision cell across the full m/z precursor range in subsequent experiments. In the LC/MSE DIA approach, alternating MS scans are acquired at low or elevated collision energies, which results in alternate MS scans of precursor and product ion spectra for all precursors in parallel (32–34). In each DIA approach, product ion spectra are reconstructed from the complex fragmentation datasets for subsequent searches against protein sequence databases or library spectral databases utilizing appropriate software.
In addition to improvements in sensitivity and proteome coverage, recent advances in MS-based quantification strategies now allow for both the relative and absolute quantification of proteins from complex proteomic mixtures. Although relative quantification using either label-free (35) or isotope labelling (36–39) methodologies have been prevalent for the past 15 years for non-hypothesis driven proteomics experiments, these approaches only measure relative protein abundance differences between two biological “states”. More recently, a number of approaches have been employed enabling the quantification of absolute molar amounts of one or more proteins within a sample. For example, multiple reaction monitoring (MRM) utilizing triple-quadrupole MS systems, an approach utilized extensively in small molecule quantification in complex sample matrices, has been applied in conjunction with isotopically labeled peptide standards to quantify selected target proteins of interest in complex biological samples (40). Unfortunately, this hypothesis-driven approach requires that one knows beforehand which protein(s) are of interest and has in hand synthetic peptide standards corresponding to these targeted proteins (41). For discovery, non-hypothesis driven experiments where absolute quantification is required across all proteins present in a sample, computational approaches have been employed which normalize MS protein signal intensity against the number of observable peptides (42, 43). These include the Exponentially Modified Protein Abundance Index (emPAI) (42) and the Intensity-Based Absolute Quantification (iBAQ) (43) approaches. Similarly, absolute molar amounts of proteins may be measured using the “Top 3” method reported with LC/MSE (34). In this approach, the average signal response of the 3 most intense peptides to any given protein per mole of protein is a constant, independent of the identity or sequence of the protein, to within +/− 10%. When complex proteomics samples are spiked with a protein standard at a known concentration and analyzed using the “Top 3” LC/MSE approach, both absolute mass (gram) and molar amounts of each identified protein may be determined relative to this standard, enabling both between and within sample mass and molar protein ratio and stoichiometry determination (34).
In this report, we have utilized the Top 3 LC/MSE approach to analyze ciliary axonemes isolated from well-differentiated cultures of human bronchial epithelial cells. Absolute quantification (molar and mass) was obtained for over 400 proteins (Supporting Information, Table SI-1), along with the relative abundances of these proteins within the axoneme, including many that have not previously been characterized as part of the ciliary proteome. Several of the previously uncharacterized proteins identified were present at relatively high abundance and were selected for further studies. RT-PCR data demonstrated that these proteins were specifically expressed during ciliated cell differentiation, and immunoblotting of isolated cilia confirmed their presence in the axoneme. Confocal immunofluorescence studies revealed unique axonemal locations for the uncharacterized proteins, further demonstrating the complexity of this unique organelle.
Experimental Procedures
Cell Culture, Cilia Isolation and Protein Extraction
Human airway epithelial (HAE) cells from non-smoking donors were obtained from the UNC Tissue Procurement and Cell Culture Core under protocols approved by the Institutional Review Board for the Protection of the Rights of Human subjects at UNC and cultured as previously described in detail (44, 45). Briefly, ~5×106 undifferentiated HAE cells were cultured in 10 cm plastic dishes for 3 days in BEGM media supplemented with penicillin (100 U/mL), streptomycin (100 µg/mL), For obtaining differentiated cells, the HAE cells were cultured in ALI media supplemented with penicillin (100 U/mL) and streptomycin (100 µg/mL) at an air/liquid interface on collagen-coated culture inserts (Millicell, Millipore) until they were heavily ciliated (greater than 4 weeks). Protein extracts from undifferentiated and differentiated airway cells were obtained using RIPA buffer (ThermoFisher Scientific) supplemented with protease inhibitors (Protease Inhibitor Cocktail, Sigma P8340). Ciliary axonemes were isolated from heavily ciliated cultures as previously described (18). Briefly, cultures were washed with phosphate-buffered saline (PBS) to remove mucus and cell debris. Deciliation buffer (10 mM Tris pH 7.5, 50 mM NaCl, 10 mM CaCl2, 1 mM EDTA, 0.1% Triton X-100, 7 mM β-mercaptoethanol, 1% protease inhibitor cocktail) was added to the surface and the culture was rocked gently for 1 min. The supernatant was collected, and the procedure was repeated. The two washings were pooled and after pelleting debris at 1000 × g, the ciliary axonemes were collected by centrifugation at 16,000 × g. Ciliary pellets were resuspended in 2× LDS buffer (Invitrogen) for immunoblot or for proteomics analysis as described (18).
Immunoblotting
The protein concentration of differentiated and undifferentiated human airway cell lysates was determined using the BCA method (Pierce BCA kit, ThermoFisher Scientific). From one 30 mm Millicell ~70 µg of ciliary protein was obtained. In order to have comparable signals between samples, 1/10th of the cilia extract (~ 7 µg of total protein) and ~1/20th of the total cell lysates were loaded. The samples were electrophoresed on NuPage 4 – 12% Bis-Tris gels in MES-SDS running buffer (ThermoFisher Scientific) and transferred to 0.45 um nitrocellulose membrane for immunoblotting. The antibodies and dilutions used are listed in Table SI-2.
Immunofluorescence
Fluorescent immunocytochemistry was performed on isolated HAE cells. To ensure the adhesion of ciliated cells to the glass, slides used for confocal imaging and cover slips used for super resolution imaging were pretreated with 1% Alcian Blue (Sigma) aqueous. The surface of the slide/cover slip was covered with the Alcian Blue solution and incubated for 20 min, then thoroughly rinsed with distilled water, until the water ran clear. The slides/cover slips were maintained in an ice cold closed chamber. The apical region of the ciliated HAE culture was washed three times for 5 min in PBS to remove mucus and debris. The media was aspirated and the basolateral and apical sides of the cultures were washed twice for 1 min in PBS. The cells were scraped out of the dish using a pipette tip, resupsended in PBS and added to the Alcian-Blue-pretreated slide/cover slip. The cells were allowed to settle for 20 min in the cold chamber and subsequently fixed for 10 min with ice cold 2% PFA or methanol. After 2 washes with ice cold PBS some slides were treated with 0.1% Triton X-100 in PBS for 5 min. For antibody staining, all the samples were processed as follows: after 2 washes in PBS, samples were incubated in blocking solution (PBS plus 10% fetal bovine serum, 0.1% Triton X-100, and 3% bovine serum albumin) for 1 h at room temperature. Primary antibodies (see Supporting Information, Table SI-2) were diluted in blocking solution and incubated with samples overnight at 4°C. After 3 washes in washing solution (PBS plus 1% fetal bovine serum, 0.1% Triton X-100, and 3% bovine serum albumin), fluorescent secondary RRX-conjugated (1:500 dilution; Jackson ImmunoResearch) or Alexa 647- or Alexa 488-conjugated (1:500 dilution; Molecular Probes Inc.) antibodies were applied for 2 h at room temperature. To stain nuclei, Hoechst 33342 (Invitrogen) was used. Samples were mounted in ProLong Gold (Invitrogen) and imaged using a Leica SP8 inverted confocal or embedded in MEA (100 mM β-Mercaptoethylamine, Sigma 30070, in PBS pH7.4) for Ground State Depletion (GSD) Super Resolution microscopy (Leica).
Immunofluorescence Quantification and Image Processing
Immunocytochemistry on cells isolated from at least 3 different donors was performed as described above for the detection and localization of proteins of interest. Samples were co-stained with an antibody detecting Ac-TUB to visualize cilia. Micrographs from different fields were taken with a Leica SP8 inverted confocal microscope using a HC PL APO 63×/1.40 Oil CS2 objective and optical zoom of 1 for quantification of positive labeled cells and fluorescence intensity, or with an optical zoom of 3 for image processing. Thus, the size of the scan field was 238 × 238 µm or 79.3 × 79.3 µm, respectively. The sample areas were scanned at a resolution of 1024*1024 pixels (pixel size of 0.23 µm with optical zoom of 1 and pixel size of 0.077 µm with optical zoom of 3). The quantification of positive cells was performed using the Cell Counter plugin of ImageJ (46). The fluorescence intensities of the different segments of the cilia were determine using Fiji (47), adapted from previously described methods (48, 49). Briefly, the cilia length was measured and then divided in 3 regions of interest (ROI), proximal (half of the length of the cilia) middle (quarter of the length of the cilia) and tip (the remaining quarter of the cilia length) (Fig. SI-1). Using the ROI manager plugin, the area, the mean gray value and the integrated density (IntDen) of each ROI as well as for 4 to 5 ROI surrounding the cells (background, Bg) were measured; 4–9 cells were measured for each protein. The corrected total cilia fluorescence (CTCF) in each ROI was calculated using the formula
The calculated fluorescence intensities for each channel (protein of interest) and Ac-TUB were plotted using GraphPad Prism version 7.1. Confocal images were converted to digital images using Photoshop at a resolution of 300dpi.
RNA Expression
Human airway epithelial cells were plated on collagen-coated inserts for culture at the air/liquid interface as described above. Under these conditions the cells initially grow as a mono-layer of undifferentiated cells. The cells begin to differentiate and after 8 to 14 days ciliated cells start to appear and subsequently increase in number. From these cultures, total RNA was isolated after 4 to 35 days of culture using an RNeasy kit (Qiagen Inc.) according to the manufacturer's protocols. Semi-quantitative RT-PCR was performed using standard procedures to determine the expression levels of transcripts of the proteins of interest. The primers used are listed in Table SI-3.
Proteomic Analysis
Axonemal pellets isolated from heavily ciliated cultures as described above were resuspended in SDS sample buffer and loaded onto 4–12% Nu-PAGE gradient gels (Novex) for separation. Axonemes from three different cell donors were analyzed independently. Following staining with Coomassie Blue, the entire lane from each replicate was cut into 18 sections, the width of the section dependent upon the intensity of protein bands present in that region of the gel, with the goal of equalizing to the extent possible the amount of protein contained in each band (Fig. SI-2). In most cases, multiple visible bands were contained within a given section, with the exception being the thick, intense band corresponding to highly abundant α- and β–tubulin at approximately 50 kDa. Each section was subjected to in-gel trypsinization using the protocol of Wilm et al. (50), and a predigested BSA standard (MassPrep Standard, Waters Corporation) was spiked into each digested band to provide a level of 250 fmol BSA per injection.
Mass Spectrometry Analysis and Database Searching
In-gel digests were analyzed by LC/MSE using a NanoAcquity UPLC and Q-Tof Premier mass spectrometry system (Waters Corporation) as previously described (51). LC/MSE raw data files were processed for database searching using ProteinLynx Global Server v2.4 (PLGS) (Waters), and subsequently searched against the human subset of the Uniprot database (Uniprot.org) using the integrated IDENTITYE search algorithm included in PLGS software package. The human database downloaded in September 2012 contained 22,631 entries, and the sequence for BSA was added to the database to enable identification of the spiked BSA internal standard protein. Default database search engine parameters were used as follows: Precursor Mass Tolerance-Automatic (0.01 Da for first pass); Product Mass Tolerance-Automatic (0.025 Th for first pass); Minimum Fragment Ion Matches per Peptide-3; Minimum Fragment Ion Matches per Protein-7; Minimum Peptide Matches per Protein-1; Maximum Protein Mass-1×106; Missed Cleavages-1; Fixed Modifications-Carbamidomethyl Cys; Variable Modifications- Amidation-C terminus; Oxidation-Met; Phosphorylation-Ser, Thr, Tyr; PLGS Set FDR-100%; Calibration Protein-BSA; Calibration Protein Concentration-250 fM. The FDR was set to 100% for the IDENTITYE search as recommended by Proteome Software for subsequent data loading into their Scaffold software in order to provide a sufficient number of incorrect peptide spectral matches (PSMs) to obtain appropriate statistics from the Peptide and Protein Prophet rescoring algorithms within Scaffold.
Data Analysis
IDENTITYE database search results were loaded for further analysis into Scaffold v4 (Proteome Software), with all 18 bands corresponding to each biological replicate loaded into a Scaffold Biosample corresponding to each replicate. Data were filtered to only display proteins meeting the following criteria: Protein Prophet threshold-95%; Peptide Prophet threshold-80%; Minimum number peptide matches-2. With these Scaffold filter parameters, the Protein Prophet FDR reported by Scaffold for the dataset was 0.1% while the Peptide Prophet FDR was reported to be 4.7%. The dataset with molar (fmol) amounts reported for each detected protein was exported into Excel for further analysis. Data from all three replicates was included in the qualitative analysis of proteins present in the axonemal preparations. In one of the replicates, an increased amount of protein was analyzed in an attempt to identify additional low abundance proteins. While additional proteins were identified, the different mass amounts obtained prevented the inclusion of this replicate for determination of protein amounts, except in the case of the radial spoke proteins (see results). In Excel, the reported fmol amounts obtained from two replicates were utilized in conjunction with the reported molecular weight for each observed protein to calculate the mass amounts (ng) of each observed protein. The total amount of protein present per replicate was estimated by summing the reported ng amounts for all identified proteins.
Post-proteomic Analysis
To investigate whether the proteins identified have previously been reported to be present in ciliary axonemes, we first searched the PubMed database using the gene name. If a previous report identified the protein as a component of cilia or flagella, the protein was considered “known” and a representative reference is included in Table SI-1. If a published reference existed, but only demonstrated expression at the RNA level, the protein was considered a “likely” axonemal component. Proteins with published references demonstrating non-axonemal expression were classified as other (i.e., contaminants). For proteins with no specific published references in PubMed, we searched the Gene section of PubMed and the Universal Protein Resource (UniProt) databases for evidence of axonemal localization or function. Proteins not classified by these methods were searched against the Cildb (V3.0; Arnaiz et al., Cilia, 2014) database. Proteins for which no published evidence of axonemal localization was found at the time of analysis were classified as unknown. Although the assignment of proteins to a specific category was current at the time of submission, during the course of these studies, several previously uncharacterized proteins have been identified as axonemal components.
Results
To identify additional components of human respiratory cilia and to begin to examine their relative abundance, ciliary axonemes were isolated from well-differentiated cultures of human airway epithelial cells by detergent extraction and differential centrifugation. As shown previously, this method yields a highly enriched preparation of ciliary axonemes (18). Axonemes were isolated from cells obtained from three different individuals and analyzed separately to provide biological replication. Axonemal proteins were separated by gradient gel electrophoresis, the gel lane was cut into 18 individual “bands” or sections (Fig. SI-2), and in-gel tryptic digests of each section were analyzed by LC/MSE as detailed in Experimental Methods.
Protein Identification and Quantification
Combining the results of these three biological replicates resulted in the identification and semi-quantification of over 400 unique proteins with at least two matching peptides at a protein FDR of 0.1% and peptide FDR of 4.7% (Table SI-1). Of these identified proteins, 352, or 88%, were observed in at least 2 of the 3 biological replicate samples. As expected, examination of the two replicates used for quantification revealed that proteins observed in only one replicate tended to be present at lower levels than those observed in both replicates. For example, the average molar amount (fmol per sample) for proteins observed in only one of the two quantified replicates was 74 fmols (median value 60 fmol), whereas for those proteins observed in both replicates, the average molar amount detected was 642 fmol (median value 170 fmol (replicate 1)). This suggests that proteins observed in only one of the three replicates may have been near or below the limit of detection for the LC/MSE acquisition methodology employed. Reasons for this could include biological variability in axonemal protein expression levels for some proteins or splitting the gel “band” for a given protein between adjacent sections during gel cutting. It was noted that several proteins previously reported to be present in airway cilia were observed in only one of the three replicates. For example, Cilia- and Flagella-Associated Protein 221 (CFAP221, (52)) and A-Kinase Anchor Protein 14 (AKAP14, (53)) and are both well-known components of human airway cilia but were only observed in one of the two quantified replicates. For this reason, we retained and report here proteins identified in only one or both replicates so as not to inadvertently exclude potential bona fide ciliary axonemal proteins that might be present at low molar amounts in our replicate samples.
Using the Top 3 absolute quantification approach with LC/MSE as described by Silva et al. (34), we were able to semi-quantify absolute amounts (fmol and ng quantities) of 401 proteins observed in these biological replicate samples (Table SI-1). We view our analysis as semi-quantitative rather than quantitative due to several reasons relating either to caveats of the LC/MSE methodology or sample related factors. It has been noted that the magnitude of quantification errors observed with LC/MSE relies on the ability to “correctly identify the top three most intense tryptic peptides”, with larger errors associated with smaller proteins (34). With our samples, as shown in Figure SI-2, there is a large range of protein abundances observed within the ciliary axonemes. For example tubulin, shown as band 10, is by far the most abundant protein in the ciliary axonemes, and quantitative measurements for tubulin and other highly abundant proteins are likely to be biased towards lower values for that reason, as they are well above the linear range for this method. Similarly, values assigned for extremely low abundance proteins are likely to be biased as well. Another issue contributing to non-ideal quantification with the Top 3 LC/MSE approach relates to the protein inference problem (54). For proteins sharing extensive sequence identity such as α- and β-tubulin and their isoforms (see below), detection of an isoform or variant specific peptide is required to unambiguously identify and assign a specific isoform. However, in these cases, one or more of the “Top 3” peptides used for quantification will likely be shared among the multiple isoforms, precluding accurate apportioning of amounts among the individual isoforms. The dynein heavy chains are another group of highly homologous proteins that also suffer from this limitation. Most importantly for our purposes, the semi-quantification afforded by LC/MSE in our study provided a way to reasonably estimate the relative amounts of identified proteins on a mass basis (% by mass) in broad terms (low, medium or highly abundant), thereby providing a means of prioritizing interesting, highly abundant proteins for further biological verification.
As expected, many of the proteins identified have been previously reported as components of cilia or ciliated cells. Searching of publicly available databases (see Experimental Procedures) revealed 205 proteins with a published reference definitively supporting their identification as part of the ciliary proteome (Table SI-1). Another 65 proteins have genetic data supporting their assignment to the ciliary transcriptome, and therefore are also likely components of the ciliary axoneme. Together, this indicates that most of the proteins identified are axonemal proteins (~70%, Fig. 1), and our data provides independent evidence for many proteins previously identified by only genetic studies. Removing proteins that were likely contaminants (mitochondrial, ribosomal, nuclear, etc.) from the remaining proteins, resulted in a group of 39 (~10%), mostly uncharacterized, proteins with no evidence regarding their presence in cilia.
Figure 1.
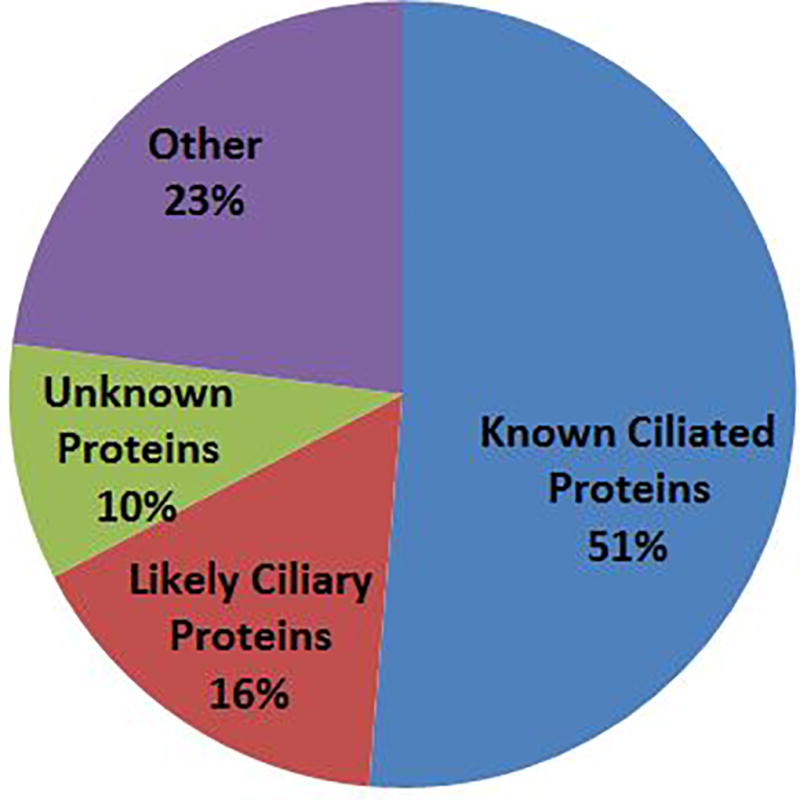
Classification of proteins identified by MS in this study. The majority of the proteins (67%) are known or likely axonemal proteins; about 10% of the proteins are previously uncharacterized proteins that are candidate axonemal components.
Tubulins and Dyneins
Of the known ciliary proteins, tubulin would be expected to be the most abundant protein identified. As expected, the predominant proteins identified in the 50 kDa section of the gel were indeed α- and β-tubulin. Manual inspection of the data identified 7 unique isoforms of α-tubulin and 13 unique isoforms of β-tubulin based on the identification of peptides exclusive to these isoforms (Table 1; see also Table SI-4a and SI-4b). The surprisingly large number of tubulin isoforms identified is in contrast to earlier, antibody based investigations (55, 56), and again points to the complex structure of the axoneme. However, because the quantity of tubulin was so large, and because of the protein inference problem with multiple isoforms of tubulin, we were unable to estimate the relative amount of the different tubulins identified.
Table 1.
Tubulin isoforms identified by LC/MSE in ciliary axonemes. The number of unique peptides from each isoform is shown.
| Accession Number | Alpha tubulin | Number of Exclusive Peptides |
|---|---|---|
| * TBA8_HUMAN | Tubulin alpha-8 chain OS=Homo sapiens GN=TUBA8 PE=1 SV=1 | 7 |
| TBA4A_HUMAN | Tubulin alpha-4A chain OS=Homo sapiens GN=TUBA4A PE=1 SV=1 | 6 |
| TBA4B_HUMAN | Putative tubulin-like protein alpha-4B OS=Homo sapiens GN=TUBA4B PE=5 SV=2 | 5 |
| * TBA3E_HUMAN | Tubulin alpha-3E chain OS=Homo sapiens GN=TUBA3E PE=1 SV=2 | 2 |
| * TBAL3_HUMAN | Tubulin alpha chain-like 3 OS=Homo sapiens GN=TUBAL3 PE=1 SV=2 | 2 |
| * TBA3C_HUMAN | Tubulin alpha-3C/D chain OS=Homo sapiens GN=TUBA3C PE=1 SV=3 | 1 |
| * TBA1C_HUMAN | Tubulin alpha-1C chain OS=Homo sapiens GN=TUBA1C PE=1 SV=1 | 1 |
| Beta Tubulin | ||
|
| ||
| TBB8_HUMAN | Tubulin beta-8 chain OS=Homo sapiens GN=TUBB8 PE=1 SV=2 | 8 |
| TBB6_HUMAN | Tubulin beta-6 chain OS=Homo sapiens GN=TUBB6 PE=1 SV=1 | 8 |
| TBB1_HUMAN | Tubulin beta-1 chain OS=Homo sapiens GN=TUBB1 PE=1 SV=1 | 8 |
| TBB5_HUMAN | Tubulin beta chain OS=Homo sapiens GN=TUBB PE=1 SV=2 | 7 |
| TBB3_HUMAN | Tubulin beta-3 chain OS=Homo sapiens GN=TUBB3 PE=1 SV=2 | 5 |
| TBB8L_HUMAN | Tubulin beta-8 chain-like protein LOC260334 OS=Homo sapiens PE=1 SV=1 | 4 |
| * YI016_HUMAN | Putative tubulin beta chain-like protein ENSP00000290377 OS=Homo sapiens PE=5 SV=2 | 3 |
| TBB4Q_HUMAN | Putative tubulin beta 4q chain OS Homo sapiens GN TUBB4Q PE 5 SV 1 | 3 |
| TBB4B_HUMAN | Tubulin beta-4B chain OS=Homo sapiens GN=TUBB4B PE=1 SV=1 | 2 |
| TBB4A_HUMAN | Tubulin beta-4A chain OS=Homo sapiens GN=TUBB4A PE=1 SV=2 | 2 |
| * TBB2A_HUMAN | Tubulin beta-2A chain OS=Homo sapiens GN=TUBB2A PE=1 SV=1 | 2 |
| TBB2B_HUMAN | Tubulin beta-2B chain OS=Homo sapiens GN=TUBB2B PE=1 SV=1 | 1 |
| * TBB4_HUMAN | Tubulin beta 4 chain OS Homo sapiens GN TUBB4 PE 1 SV 2 | 1 |
These particular isoforms did meet the stringency threshold for inclusion in the full proteomic dataset (95% Protein Probability, 80% Peptide Probability, 2 Peptide Minimum). They were nonetheless included in this list of isoforms due to the evidence for their presence in the axoneme. The complete list of tubulin peptides recovered is shown in Table SI-4.
Dynein heavy chains (DHC) were also detected at high abundances. In all, twelve distinct DHCs were identified on the basis of detected peptides exclusive to each DHC (Table 2; see also Table SI-5), and most of these have previously been reported. However, our data provides the first biochemical evidence for the presence of DHC 2, 6, and 12. Similar to the highly conserved tubulin isoforms, the DHCs are also highly conserved, especially in their catalytic domains, precluding our ability to assign relative abundances to the various DHCs.
Table 2.
Dynein heavy chains identified by LC/MSE in ciliary axonemes. The number of unique peptides from each isoform is shown. All proteins were detected in both replicates.
| Accession Number | Exclusive To |
1Number of Exclusive Peptides |
Molecular Weight (kDa) |
|---|---|---|---|
| DYH9_HUMAN | Dynein heavy chain 9, axonemal OS=Homo sapiens GN=DNAH9 PE=1 SV=3 | 198 | 512 |
| * DYH8_HUMAN | Dynein heavy chain 8, axonemal OS=Homo sapiens GN=DNAH8 PE=1 SV=2 | 9 | 515 |
| DYH7_HUMAN | Dynein heavy chain 7, axonemal OS=Homo sapiens GN=DNAH7 PE=1 SV=2 | 98 | 461 |
| DYH6_HUMAN | Dynein heavy chain 6, axonemal OS=Homo sapiens GN=DNAH6 PE=1 SV=3 | 118 | 476 |
| DYH5_HUMAN | Dynein heavy chain 5, axonemal OS=Homo sapiens GN=DNAH5 PE=1 SV=3 | 245 | 529 |
| * DYH3_HUMAN | Dynein heavy chain 3, axonemal OS=Homo sapiens GN=DNAH3 PE=2 SV=1 | 2 | 471 |
| DYH2_HUMAN | Dynein heavy chain 2, axonemal OS=Homo sapiens GN=DNAH2 PE=1 SV=3 | 58 | 508 |
| DYH17_HUMAN | Dynein heavy chain 17, axonemal OS=Homo sapiens GN=DNAH17 PE=2 SV=2 | 7 | 512 |
| DYH12_HUMAN | Dynein heavy chain 12, axonemal OS=Homo sapiens GN=DNAH12 PE=1 SV=2 | 74 | 357 |
| DYH11_HUMAN | Dynein heavy chain 11, axonemal OS=Homo sapiens GN=DNAH11 PE=1 SV=3 | 120 | 521 |
| DYH10_HUMAN | Dynein heavy chain 10, axonemal OS=Homo sapiens GN=DNAH10 PE=1 SV=4 | 63 | 515 |
| DYH1_HUMAN | Dynein heavy chain 1, axonemal OS=Homo sapiens GN=DNAH1 PE=1 SV=4 | 117 | 494 |
Number of exclusive peptides in replicate with most exclusive peptide matches.
These particular isoforms did meet the stringency threshold for inclusion in the full proteomic dataset (95% Protein Probability, 80% Peptide Probability, 2 Peptide Minimum). They were nonetheless included in this list of isoforms due to the evidence for their presence in the axoneme. The complete list of dynein heavy chain peptides recovered is shown in Table SI-5.
Radial Spoke Proteins
Nine proteins that are known components of the radial spoke assembly were identified, including RSPH1, RSPH3, RSPH4A, RSPH9, ROP1L, PPIL6, RTDR1, DNAJB13, and NME5. These proteins accounted for ~1% of the total axonemal mass. Table 3 shows the relative amounts of the radial spoke proteins from the three analysis, calculated by dividing the molar amount of each protein detected by the molar amount of the least abundant radial spoke protein in that sample. Although there was some variability between experiments (see File SI-2), the data clearly indicates that RSPH9 and RSPH4A were the most abundant radial spoke proteins identified, and were 2–3 fold more abundant on a molar basis than RSPH1 and RSPH3 in our samples. RTDR1, PPIL6, and NME5 were present at lower abundance. While several elegant studies (e.g., 14, 57, 58) have begun to determine the detailed molecular structure and composition of the radial spokes in other species, little is known about the relative abundance of individual radial spoke proteins in human cilia. Comparing the predicted stoichiometry of these proteins with those from a published model of a C. reinhardtii radial spoke (57) (Table 3) shows a similar pattern of abundance, with the spoke head proteins generally more abundant than those of the stalk or neck. However it should be noted that while axonemes from C. reinhardtii contain two RS that are believed to be identical, our data represents the average of (at least) two different RS structures. Additional studies will need to be performed to determine the exact composition of the individual human radial spokes.
Table 3.
Relative molar amount of radial spoke components determined from 3 separate axonemal isolates. The amount of each protein was determined by LC/MSE and normalized to the radial spoke protein with the lowest abundance: the average relative amount is shown (see also supplemental figure S#. The estimated stoichiometry is compared to the estimated stoichiometry of Chlamydomomas, as reported in Pigino et al., 2011 (57).
| Identified Proteins | Accession Number |
Molecular Weight |
Average Relative Amount |
Estimated Stoichiometry |
Chlamydomonas reinhardtii |
Estimated Stoichiometry$ |
|---|---|---|---|---|---|---|
| Radial spoke head protein 4 homolog A | RSH4A_HUMAN | 81 kDa | 11.6 | 12 | RSPH4A | 4 |
| Radial spoke head protein 9 homolog | RSPH9_HUMAN | 31 kDa | 7.8 | 8 | RSPH9 | 4 |
| Radial spoke head 1 homolog | RSPH1_HUMAN | 35 kDa | 5.8 | 6 | RSPH1 | 4 |
| DnaJ (Hsp40) related, subfamily B, member 13 | B3LEP4_HUMAN | 36 kDa | 5.2 | 5 | RSP16 | 2 |
| Radial spoke head protein 3 homolog | RSPH3_HUMAN | 64 kDa | 4.2 | 4 | RSPH3 | 4 |
| Ropporin-1-like protein | ROP1L_HUMAN | 26 kDa | 3.5 | 4 | RSP11 | 2 |
| Rhabdoid tumor deletion region protein 1 | RTDR1_HUMAN | 39 kDa | 2.1 | 2 | RSP14 | 1 |
| Nucleoside diphosphate kinase homolog 5 | NDK5_HUMAN | 24 kDa | 1.9 | 2 | RSP23 | 2 |
| Peptidyl-prolyl cis-trans isomerase-like 6 | PPIL6_HUMAN | 35 kDa | 1.6 | 2 | RSP12 | 2 |
Uncharacterized Proteins
In addition to identifying many previously known components of the ciliary proteome, these studies identified ~40 proteins that were not previously reported to be present in the human ciliary proteome at the time of the original analyses (Table SI-1). Many of these proteins were uncharacterized, with little information available as to their function. However, several of them are present in the proteome at substantial amounts. For example, ERICH3 accounts for over 1% of the total axonemal mass: together these proteins may account for ~10% of the axonemal proteome (i.e., ~40 out of 400). To confirm these proteins are present in the axoneme and to begin to characterize their expression and localization, we investigated eight of the most abundant uncharacterized proteins (ERICH3, C1orf87, CFAP77, SPACA9, CCDC181, CCDC173, CFAP61, and CCDC17; Table 4) further.
Table 4.
Uncharacterized proteins identified by LC/MSE and selected for further study. All of these proteins were of high relative abundance, as determined by the top 3 peptides identified.
| Current Uniprot Name | Gene | Synonym | Average Number of Exclusive Peptides |
Molecular Weight (kDa) |
Replicate A Amount (ng) |
Replicate B Amount (ng) |
Average Amount (ng) |
Average Mass Percentage |
|---|---|---|---|---|---|---|---|---|
| Glutamate-rich protein 3 | ERICH3 | C1orf173 | 102 | 168 | 554 | 511 | 533 | 1.7% |
| Uncharacterized protein C1orf87 | C1orf87 | 33 | 62 | 115 | 67 | 91 | 0.2% | |
| Cilia- and flagella-associated protein 77 | CFAP77 | C9orf171 | 15 | 36 | 43 | 65 | 54 | 0.2% |
| Sperm acrosome-associated protein 9 | SPACA9 | C9orf9 | 11 | 25 | 16 | 72 | 44 | 0.2% |
| Coiled-coil domain-containing protein 181 | CCDC181 | C1orf114 | 12 | 60 | 36 | 30 | 33 | 0.1% |
| Coiled-coil domain-containing protein 173 | CCDC173 | C2orf77 | 45 | 66 | 25 | 33 | 29 | 0.1% |
| Cilia- and flagella-associated protein 61 | CFAP61 | C20orf26 | 16 | 141 | 15 | 37 | 26 | 0.1% |
| Coiled-coil domain-containing protein 17 | CCDC17 | 20 | 68 | 24 | 20 | 22 | 0.1% |
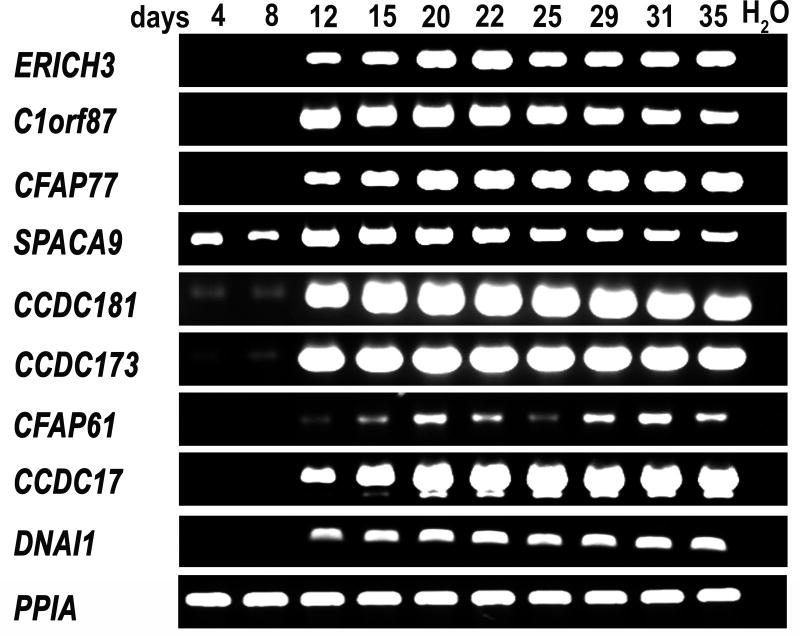
We first examined the transcript levels of the uncharacterized proteins during ciliated cell differentiation. Human airway epithelial cells were plated on collagen coated membranes and allowed to differentiate at the air/liquid interface. As described above, these cultures initially consist of a layer of undifferentiated basal-like cells that begin to express ciliated cell markers after approximately 2 weeks of culture and become heavily ciliated after 3–4 weeks. Using RT-PCR with specific intron-spanning primers, we analyzed the relative levels of each protein’s transcript (RNA abundance) at different time points of differentiation (Fig. 2). The transcripts for 6 of the 8 uncharacterized proteins (ERICH3, CFAP77, C1orf87, CCDC181, CCDC173, and CCDC17 were barely detectable at the early time points (days 4 and 8) when the cultures were mostly undifferentiated, while all 6 of these transcripts were strongly present at the later stages of culture (days 12 thru 35) when ciliated cells became abundant. SPACA9 was expressed at detectable levels in the early cultures, but its expression level also increased as the cultures underwent differentiation. CFA61 appeared to be expressed at a lower level than the other proteins, but followed a similar pattern of expression, with transcripts undetectable at days 4 and 8, weakly detectable at day 12, and present at variable levels at later time points. Overall, the pattern of expression of these uncharacterized proteins is consistent with that observed for other known axonemal proteins (e.g., DNAI1 (59)).
Figure 2.
RT-PCR analysis of uncharacterized proteins during differentiation of human airway epithelial cells in vitro. RNA was isolated from air/liquid interface cultures on the indicated days and specific primers for each of the proteins were used to analyze relative expression level. Expression of all of the uncharacterized proteins increased during differentiation. The known ciliary component, dynein axonemal intermediate chain 1 (DNAI1) was amplified as a positive control, while cyclophilin A (PPIA) was amplified as a loading control; reactions with no RNA (H2O) were used as a negative control.
We then used immunoblotting to determine if the uncharacterized proteins were detectable in differentiated cells, and more specifically, in the isolated ciliary axoneme (Fig. 3). None of the proteins were detected in the undifferentiated cell lysate. Most of the proteins (5 out of 8) were clearly detected in the differentiated cell lysate, whereas SPACA9 was weakly detected in the cell lysate, and CFAP77 and CFAP61 were undetectable. In contrast, all of the proteins were clearly present in the isolated axonemal preparation. These results support the conclusion that these uncharacterized proteins are integral components of the ciliary axoneme.
Figure 3.
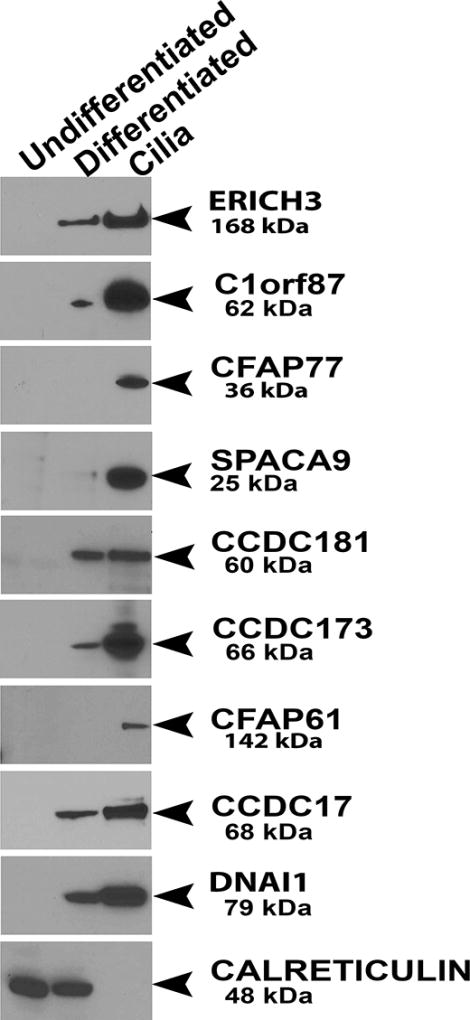
Western blot showing the expression of the uncharacterized proteins in whole cell lysates from undifferentiated (3 days) and differentiated (greater than 4 weeks) HBE cells and isolated ciliary axonemes. All proteins were undetectable in the undifferentiated cells but were clearly present in the isolated axonemes. Samples were also probed for the known ciliary component DNAI1 as a positive control, while CALRETICULIN was used as a loading control.
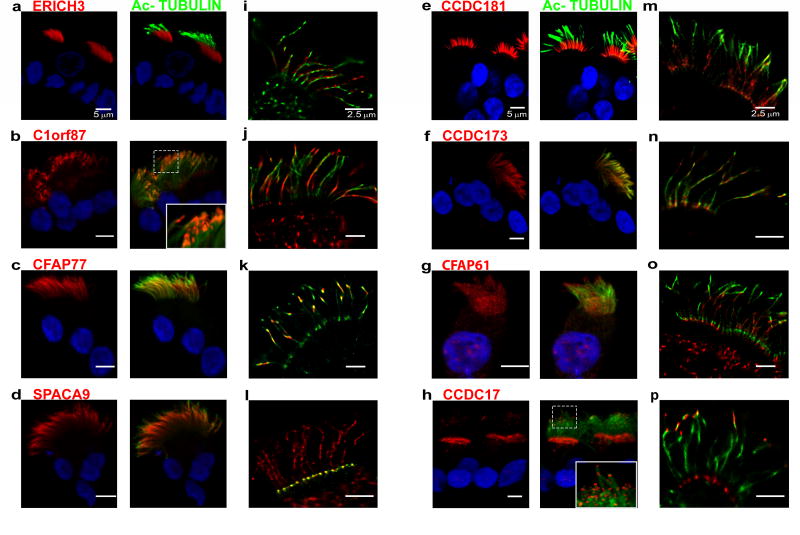
To directly confirm the ciliary localization of the eight proteins, a combination of confocal and GSD super-resolution immunofluorescent microscopy was used (Fig. 4). The number of positive ciliated and non-ciliated cells was quantified. In all cases, positive staining for these uncharacterized proteins was found exclusively in ciliated cells (Table SI-6). The staining using specific antibodies against each of the different proteins exhibited a unique pattern of staining. This was quantified by measuring the fluorescence intensity of the staining in 3 segments of the cilia; the proximal half, the middle quarter, and tip of the cilia (see methods, Fig. SI-1 and Fig. SI-3). Antibodies against ERICH3 strongly stained the proximal and middle region of the cilia (Figure 4a, 4i, and Fig. SI-3a). ERICH3 (glutamate-rich protein 3; also known as C1orf173) encodes a protein of 1530 amino acids in length with a molecular mass of 168.4 kDa (UniprotKB: Q5RHP9). It contains two domains of unknown function, DUF4590 and DUF4543. Both DUF regions are currently uncharacterized. The function of this protein is unclear; however on a statistical-analysis-based association study the expression of ERICH3 was related to incidence of osteoporosis and colorectal cancer (60). Previous microarray analysis reported the expression of ERICH3 in the brain, eye, lung, mammary gland, muscle, pituitary gland, testis, trachea, and uterus (61). In addition, a recent study suggests that ERICH3 is associated with plasma serotonin concentrations and may have a role in selective serotonin reuptake inhibitor (SSRI) therapy treatment outcomes in patients with major depressive disorder (62).
Figure 4.
Immunofluorescence localization of the indicated proteins in isolated ciliated cells (1st column; red) co-localized with Ac-TUB (2nd column; green) by conventional confocal microscopy. The 3rd column shows localization of the proteins in micrographs obtained using GSD microscopy. See text for details. Scale bar = 5 µm in left and center panels; nuclear staining was with Hoechst 33342 (blue). Scale bar = 2.5 µm in right panels.
The immunofluorescence studies of C1ORF87 using confocal laser scanning showed that this protein localizes prominently to the tip region of the cilia (Fig. 4b, 4j and Fig SI-3b), but was also consistently detected in the apical region of ciliated cells. The analysis of fluorescence intensities (Fig. SI-3b) showed that the signal for this protein is present in all 3 segments of the cilia. Super resolution microscopy, confirmed these results and revealed patches of immunoreactivity throughout the axoneme. The staining for C1ORF87 illustrates the difference in resolution and sensitivity between the two types of microscopy. We conclude that C1ORF87 is present along the cilia, but is more abundant at the tip of the cilia (see insert in Fig. 4b). C1ORF87 is 546 amino acids in length with a molecular mass of 62.0 kDa (UniProtKB: Q8N0U7). Expression of this protein was previously shown to be enhanced in ciliated cells (63). The function of this protein is still unknown but, using bioinformatics and proteomics approaches C1ORF87 was characterized as an EF-hand containing, calcium-binding protein with putative transporter function and was referred to as carcinoma related EF-hand protein (CREF) because of its association to breast, lung and liver cancer (64). The localization of C1ORF87, with prominent staining at both the base and the tip of the cilia, suggests that this protein may play a role in intraflagellar transport (65–67).
CFAP77 (cilia- and flagella-associated protein 77) appears to localize along the length of the axoneme (Fig. 4c, 4k and Fig. SI-3c) but is more abundant at the middle piece of the cilia. The CFAP77 protein contains 320 amino acids and has a molecular mass of 36.5 kDa (UniProtKB: Q6ZQR2). The function of this protein is still unknown but, in a previous study using genome wide RNA sequencing and immunohistochemistry, enhanced expression of CFAP77 was detected in ciliated cells (63).
SPACA9, or sperm acrosomal-associated protein 9, is a 222 amino acid protein with a molecular mass of 25.2 kDa (UniProtKB: Q96E40). Immunostaining of ciliated airway cells shows the protein localizes along the entire length of the axoneme (Fig. 4d, 4l and Fig. SI-3d). Interestingly, the available antibody against SPACA9 only provided a clear signal when the cells where not permeabilized, suggesting this protein may not be tightly associated with the axoneme. Under these conditions, the anti-Ac-TUB antibody only labels the proximal segment of the cilia (Fig. 4i). SPACA9 was reported to be a tumor suppressor (68, 69) that possess a poly-glutamine domain (4 residues) of unknown function. This protein has previously been shown to localize in the cytoplasm of the cells where it is associated with cytoplasmic microtubules (70). In mouse caudal sperm, SPACA9 localizes to sperm head, and is also present on the midpiece and principle piece of sperm tails. This localization is regulated by Y chromosome (71).
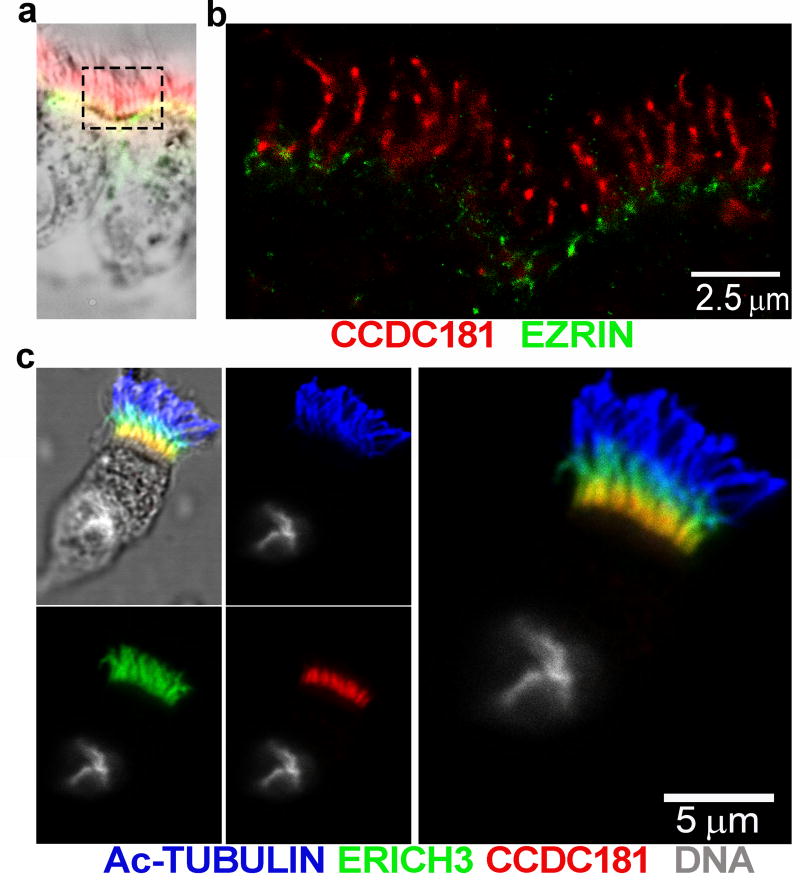
CCDC181 (also known as C1orf114) encodes a coiled-coil domain containing protein of 509 amino acids (UniProtKB: Q5TID7) that localizes to the proximal portion of the ciliary axoneme. However, unlike ERICH3, CCDC181 appears to localize to a narrower region of the proximal axoneme (Fig. 4e, 4m and Fig. SI-3e), being clearly detectable in only approximately 25% of the most proximal axoneme. Because this region also contains the microvilli, we also co-stained ciliated cells for the microvilli marker EZRIN. Microvilli are shorter (~1 µm) and narrower than cilia (~7 µm). As shown in Figure 5a and 5b, the staining for EZRIN localizes at the apical region of ciliated cells while the staining of CCDC181 extends from the cell surface and is found at the proximal region of the cilia. Because CCDC181 and ERICH3 showed a similar pattern of localization at the proximal region of the axoneme, we also labeled ciliated cells using specific antibodies for both proteins. As shown in Figure 5c, CCDC181 does co-localize with the signal for Ac-TUB, but does not completely overlap the signal for ERICH3. Considering the high versatility of the coiled-coiled protein folding motif (72), CCDC181 could be involved in almost any aspect of ciliary structure or function.
Figure 5.
a) Localization of CCDC181 (red) and ezrin (green) confirms that CCDC181 is a component of the ciliary axoneme and not the microvilli. A low magnification bright field image of the stained ciliated cells (a) imaged with super resolution microscopy (b) showing that CCDC181 localizes specifically to the cilia (scale bar = 2.5 µm). c) Co-localization of Ac-TUB (blue), ERICH3 (green), and CCDC181 (red) by immunofluorescence localizes CCDC181 to the proximal portion of airway cilia. Cell nuclei were stained with Hoechst 33342 (gray), scale bar = 5 µm.
The protein CCDC173 (also known as C2orf77) protein is 552 amino acids long with a molecular mass of 66.4 kDa (UniProtKB: Q0VF26). CCDC173 contains 4 coiled-coil domains, and localizes along the entire length of the ciliary axoneme. Nothing is known about the function of this protein; however the 4 coiled coil domains and its distribution along the axoneme (Fig. 4f, 4n and Fig. SI-3f) suggest that it forms a part of the regular repeating units of the axoneme. It therefore may be involved in protein interactions that are important for the assembly and/or mechanical stability to the axoneme.
Antibodies against CFAP61 label the length of the cilia (Fig. 4g, 4o and Fig. SI-3g); however the signal with this antibody was weaker than that observed with other proteins, e.g. SPACA9. This is likely due to the ~10-fold lower levels of CFAP61 compared to SPACA9 (184 vs 1771 fmol). CFAP61 (UniProtKB: Q8NHU2) is a large protein (114 kDa) of unknown function, although it shares a high degree of homology with the Chlamydomonas protein FAP61, an oxidoreductase protein identified as part of the calmodulin and radial spoke-associated complex that mediates regulatory signals between the radial spokes and dynein arms (73).
Immunostaining for CCDC17 (UniProtKB: Q96LX7) revealed a bi-modal staining pattern, with a strong signal at both the ciliary tip (see insert Fig. 4h) and the basal body region (Fig. 4h, 4p and Fig. SI-3h). This pattern can also be seen in cells stained for CCDC17 and γ-TUBULIN (Fig. SI-4). This 67 kDa protein contains 3 coiled coil domains but little is known about its function. However, its localization to both the ciliary tip and basal body region suggests that this protein may play a unique role in ciliary biology.
Discussion
The human respiratory cilium is a remarkable nanomachine. Composed of hundreds of unique interacting proteins, a single cilium beats continuously at a frequency of ~15 times per second in coordination with thousands of other cilia to transport mucus out of the airways and maintain a healthy pulmonary system. Much of what is known about the structure of the ciliary axoneme has been derived from model systems, most notably Chlamydomonas. The identification of mutations that cause primary ciliary dyskinesia has provided additional information on the function of over 30 specific ciliary proteins. However it is clear that our understanding of how the hundreds of proteins that make up a cilium function to provide efficient mucociliary clearance is incomplete. Proteomics provides a unique tool to not only identify the majority of proteins present in a complex sample (i.e., the axoneme), but also to provide a semi-quantitative estimate of the relative amounts of proteins present. We therefore used the technique of label-free LC/MSE to analyze isolated preparations of human ciliary axonemes.
While the sensitivity and specificity of MS approaches have recently increased dramatically, there are still several limitations to the technique as applied here. First, as with any analytical methodology, comprehensive, accurate quantification of sample components by LC/MSE is difficult for all proteins when amounts of individual components span a large dynamic range. In the case of the ciliary axoneme, measured fmol amounts of individual protein components spanned roughly 2 orders of magnitude (Table SI-1), well beyond the linear dynamic range of the method. Second, proteins in the ciliary axoneme which are present as multiple highly conserved isoforms may not be accurately quantified using the Top-3 LC/MSE quantification approach due to generation of identical proteolytic peptides from the various isoforms. Examples include the axonemal dynein heavy chain family (12 isoforms identified) and the tubulin family (20 α/β-isoforms identified). Finally, while the increased sensitivity of detection improves our ability to detect minor components of the ciliary axoneme, it also increases the possibility of detecting minor amounts of contaminating proteins. In the present study, cilia were isolated from differentiated cultures of human airway epithelial cells using a detergent and differential centrifugation. While this simple procedure results in a sample highly enriched for ciliary axonemes, it is not a pure sample. This is evidenced by the identification of nuclear, mitochondrial, and membrane proteins. However, additional purification steps would likely result in the loss of bona fide axonemal components. Therefore the identification of a protein in our sample does not prove axonemal localization and additional studies, for example localization by immunofluorescence microscopy, need to be performed to confirm the axonemal location of identified proteins.
Although it has long been recognized that α– and β–tubulin comprise the fundamental building blocks of ciliary axonemes, our proteomic analysis revealed a remarkable diversity of different tubulin isoforms. Several of these were present at low abundance, which may explain why they were not detected in previous studies. For example, using isotype specific antibodies, Jensen-Smith et al. (2003) (55) were able to detect tubulin βI and βIV, but not tubulin βII and βIII in tracheal cilia. Although the functional role of the different tubulin isoforms remains to be determined, these data demonstrate the sensitivity and specificity of our approach. The DHCs represent another highly conserved family of proteins that are present in different amounts in the ciliary axoneme. Again, while several of the more abundant DHCs have been previously localized to the axoneme by immunofluorescence studies (e.g., DNAH5, DNAH9, DNAH11, (19, 20)), the MS data provided here is the first biochemical evidence that several of the other DHCs are bona fide axonemal components. Among the other known axonemal proteins, the components of the radial spokes are of interest because they play a key role in the regulation of ciliary activity, and mutations in the radial spoke proteins are a cause of primary ciliary dyskinesia. The LC/MSE data obtained here provides a first approximation of the relative abundance of the different radial spoke proteins. Although the composition and structure of the radial spokes from Chlamydononas has been the subject of intense investigation, much less is known about the composition of the radial spokes from other species, including human. Because cilia from humans contain 3 complete radial spokes, while Chlamydomonas flagella only have two, it is not surprising that the relative abundance of the radial spoke proteins is also different (57, 74).
One of the unique proteins identified by our investigation was ERICH3, or glutamate-rich protein 3 (also known as C1orf173). This protein contains 2 domains of unknown function, and has not previously been reported to be a component of cilia, even though it comprised ~1.5% of the axonemal mass in our samples. In part, this may be because this protein has no close orthologue in Chlamydomonas, and therefore has not been identified in studies of this species. This protein is highly expressed in ciliated cells and localizes to the proximal half of the cilium. The relative abundance of this protein, and the lack of information concerning its function, highlights the need to study axonemes from different species or tissues independently.
Another protein that localizes to the proximal half of the cilium and has not been previously characterized is CCDC181, or coiled-coil domain containing protein 181. While CCDC181 localizes to the proximal region of the axoneme, its distribution is not the same as ERICH3, suggesting that these two proteins may not interact directly, and likely are part of different ciliary complexes. CCDC181 is not as abundant as ERICH3, comprising ~0.1% of the axoneme, but this uncharacterized protein is also easily detected in the ciliary axoneme, and through its coiled coil domain, possibly interacts with other ciliary proteins to assemble the axoneme. Interestingly, hypermethylation of CCDC181 is being investigated as a biomarker of prostate cancer (75).
In contrast, C1orf87 appears to localize along the cilia but is more abundant to the ciliary tip. This protein contains an EF-hand domain that may bind calcium, and has been reported to be down regulated in adenocarcinoma, relative to normal lung tissue (76). This is perhaps not surprising, as cancerous cells are not known to differentiate into ciliated cells. Thus the significance of this observation is unclear. Because of its location near or at the ciliary tip and its predicted ability to bind calcium, it is possible that C1orf87 plays a signaling role in the axoneme, or perhaps interacts with the overlying mucus layer.
Immunolocalization of SPACA9, CFAP77 and CCDC173 by confocal fluorescent microscopy suggests that these three proteins are located along the entire length of the axoneme, clearly in a different pattern than the above proteins. Our data demonstrates that the expression of these proteins increases during ciliated cell differentiation at the RNA level, and immunoblotting demonstrated a clear enrichment of the proteins in ciliary preparations. These data thus validate the identification of these proteins as axonemal components by the proteomic screen.
Immunofluorescent localization of CCDC17 revealed yet another pattern of localization, with CCDC17 staining detectable at both the basal body region and the tip of the cilia. Nothing is known about the function of this protein that contains 3 coiled coil domains, but it may play a role in assembly or stabilization of the ciliary tip and/or basal body. Alternatively, the protein may exist in two pools and be transported from the base to the tip as part of the intraflagellar transport mechanism (65–67).
CFAP61 (C20orf26) was also identified in our proteomic screen; and RT-PCR and immunoblot data clearly show that its expression is linked to ciliogenesis. CFAP61 was detected at a lower abundance in our proteomic analysis, and interestingly, our immunofluorescence data revealed a weak staining of the axoneme. This result supports the relative quantification of ciliary proteins obtained by LC/MSE in this study. Future studies, comparing the amount of ciliary proteins in samples from patients with PCD or genetically modified cell cultures (i.e., following knock down of specific proteins) may provide important information on the proteins that make up the various ciliary structures.
In summary, our LC/MSE-based label-free mass spectrometry approach allowed us to identify 205 known components of cilia, including 12 dynein heavy chains and 20 variants of tubulin, and 65 additional proteins that are likely components of human respiratory cilia. Some of these uncharacterized proteins are likely components of human respiratory cilia, while others are likely contaminants that are reproducibly present in our axonemal preparation. We selected several of these uncharacterized proteins that were present in relatively high abundance, and using a combination of techniques, we confirmed that they were indeed components of the ciliary axoneme. These proteins are present in different amounts, are of various sizes, contain a variety of known and unknown protein domains, and are targeted to specific locations within the ciliary axoneme. Yet their function in human respiratory cilia is completely unknown. The large number of uncharacterized proteins, the wide range of protein abundance, and the unique distribution of these proteins within the axoneme, suggest an even greater level of complexity of axonemal structure than previously realized. While the current study has provided additional information on the proteins that comprise the ciliary axoneme, to completely understand how a respiratory cilium functions, the role of each specific protein will have to be determined individually.
Supplementary Material
Acknowledgments
This research was funded in part by NIH grants DK065988, HL117836, and HL071798. The authors would also like to thank Dr. R. Tarran for facilitating the use of the Leica Sp8 Confocal and Leica GSD super resolution microscopes, Dr. S. H. Randell and the UNC Cell culture core for providing human airway cells, and Dr. S. Kreda for kindly providing the mouse anti-ezrin antibody.
References
- 1.Bustamante-Marin XM, Ostrowski LE. Cilia and Mucociliary Clearance. Cold Spring Harb Perspect Biol. 2016 doi: 10.1101/cshperspect.a028241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knowles MR, Boucher RC. Mucus clearance as a primary innate defense mechanism for mammalian airways. J Clin Invest. 2002;109:571–577. doi: 10.1172/JCI15217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wanner A, Salathe M, O'Riordan TG. Mucociliary clearance in the airways. Am. J. Respir. Crit. Care Med. 1996;154:1868–1902. doi: 10.1164/ajrccm.154.6.8970383. [DOI] [PubMed] [Google Scholar]
- 4.Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou JL, et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science (New York, N.Y.) 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 5.Rommens JM, Iannuzzi MC, Kerem B, Drumm ML, Melmer G, Dean M, Rozmahel R, Cole JL, Kennedy D, Hidaka N, et al. Identification of the cystic fibrosis gene: chromosome walking and jumping. Science (New York, N.Y.) 1989;245:1059–1065. doi: 10.1126/science.2772657. [DOI] [PubMed] [Google Scholar]
- 6.Boucher RC. An overview of the pathogenesis of cystic fibrosis lung disease. Adv Drug Deliv Rev. 2002;54:1359–1371. doi: 10.1016/s0169-409x(02)00144-8. [DOI] [PubMed] [Google Scholar]
- 7.Shapiro AJ, Zariwala MA, Ferkol T, Davis SD, Sagel SD, Dell SD, Rosenfeld M, Olivier KN, Milla C, Daniel SJ, Kimple AJ, Manion M, Knowles MR, Leigh MW Genetic Disorders of Mucociliary Clearance, C. Diagnosis, monitoring, and treatment of primary ciliary dyskinesia: PCD foundation consensus recommendations based on state of the art review. Pediatric pulmonology. 2016;51:115–132. doi: 10.1002/ppul.23304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olbrich H, Haffner K, Kispert A, Volkel A, Volz A, Sasmaz G, Reinhardt R, Hennig S, Lehrach H, Konietzko N, Zariwala M, Noone PG, Knowles M, Mitchison HM, Meeks M, Chung EM, Hildebrandt F, Sudbrak R, Omran H. Mutations in DNAH5 cause primary ciliary dyskinesia and randomization of left-right asymmetry. Nat Genet. 2002;30:143–144. doi: 10.1038/ng817. [DOI] [PubMed] [Google Scholar]
- 9.Zariwala MA, Leigh MW, Ceppa F, Kennedy MP, Noone PG, Carson JL, Hazucha MJ, Lori A, Horvath J, Olbrich H, Loges NT, Bridoux AM, Pennarun G, Duriez B, Escudier E, Mitchison HM, Chodhari R, Chung EM, Morgan LC, de Iongh RU, Rutland J, Pradal U, Omran H, Amselem S, Knowles MR. Mutations of DNAI1 in primary ciliary dyskinesia: evidence of founder effect in a common mutation. Am J Respir Crit Care Med. 2006;174:858–866. doi: 10.1164/rccm.200603-370OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boon M, Wallmeier J, Ma L, Loges NT, Jaspers M, Olbrich H, Dougherty GW, Raidt J, Werner C, Amirav I, Hevroni A, Abitbul R, Avital A, Soferman R, Wessels M, O'Callaghan C, Chung EM, Rutman A, Hirst RA, Moya E, Mitchison HM, Van Daele S, De Boeck K, Jorissen M, Kintner C, Cuppens H, Omran H. MCIDAS mutations result in a mucociliary clearance disorder with reduced generation of multiple motile cilia. Nat Commun. 2014;5:4418. doi: 10.1038/ncomms5418. [DOI] [PubMed] [Google Scholar]
- 11.Horani A, Druley TE, Zariwala MA, Patel AC, Levinson BT, Van Arendonk LG, Thornton KC, Giacalone JC, Albee AJ, Wilson KS, Turner EH, Nickerson DA, Shendure J, Bayly PV, Leigh MW, Knowles MR, Brody SL, Dutcher SK, Ferkol TW. Whole-exome capture and sequencing identifies HEATR2 mutation as a cause of primary ciliary dyskinesia. American Journal of Human Genetics. 2012;91:685–693. doi: 10.1016/j.ajhg.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knowles MR, Ostrowski LE, Loges NT, Hurd T, Leigh MW, Huang L, Wolf WE, Carson JL, Hazucha MJ, Yin W, Davis SD, Dell SD, Ferkol TW, Sagel SD, Olivier KN, Jahnke C, Olbrich H, Werner C, Raidt J, Wallmeier J, Pennekamp P, Dougherty GW, Hjeij R, Gee HY, Otto EA, Halbritter J, Chaki M, Diaz KA, Braun DA, Porath JD, Schueler M, Baktai G, Griese M, Turner EH, Lewis AP, Bamshad MJ, Nickerson DA, Hildebrandt F, Shendure J, Omran H, Zariwala MA. Mutations in SPAG1 Cause Primary Ciliary Dyskinesia Associated with Defective Outer and Inner Dynein Arms. American Journal of Human Genetics. 2013;93:711–720. doi: 10.1016/j.ajhg.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barber CF, Heuser T, Carbajal-Gonzalez BI, Botchkarev VV, Jr, Nicastro D. Three-dimensional structure of the radial spokes reveals heterogeneity and interactions with dyneins in Chlamydomonas flagella. Molecular biology of the cell. 2012;23:111–120. doi: 10.1091/mbc.E11-08-0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin J, Heuser T, Carbajal-Gonzalez BI, Song K, Nicastro D. The structural heterogeneity of radial spokes in cilia and flagella is conserved. Cytoskeleton. 2012;69:88–100. doi: 10.1002/cm.21000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amaral A, Castillo J, Estanyol JM, Ballesca JL, Ramalho-Santos J, Oliva R. Human sperm tail proteome suggests new endogenous metabolic pathways. Molecular & cellular proteomics : MCP. 2013;12:330–342. doi: 10.1074/mcp.M112.020552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baker MA, Naumovski N, Hetherington L, Weinberg A, Velkov T, Aitken RJ. Head and flagella subcompartmental proteomic analysis of human spermatozoa. Proteomics. 2013;13:61–74. doi: 10.1002/pmic.201200350. [DOI] [PubMed] [Google Scholar]
- 17.Liu Q, Tan G, Levenkova N, Li T, Pugh EN, Jr, Rux JJ, Speicher DW, Pierce EA. The proteome of the mouse photoreceptor sensory cilium complex. Molecular & cellular proteomics : MCP. 2007;6:1299–1317. doi: 10.1074/mcp.M700054-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ostrowski LE, Blackburn K, Radde KM, Moyer MB, Schlatzer DM, Moseley A, Boucher RC. A proteomic analysis of human cilia: identification of novel components. Mol Cell Proteomics. 2002;1:451–465. doi: 10.1074/mcp.m200037-mcp200. [DOI] [PubMed] [Google Scholar]
- 19.Fliegauf M, Olbrich H, Horvath J, Wildhaber JH, Zariwala MA, Kennedy M, Knowles MR, Omran H. Mislocalization of DNAH5 and DNAH9 in respiratory cells from patients with primary ciliary dyskinesia. Am. J. Respir. Crit. Care Med. 2005;171:1343–1349. doi: 10.1164/rccm.200411-1583OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dougherty GW, Loges NT, Klinkenbusch JA, Olbrich H, Pennekamp P, Menchen T, Raidt J, Wallmeier J, Werner C, Westermann C, Ruckert C, Mirra V, Hjeij R, Memari Y, Durbin R, Kolb-Kokocinski A, Praveen K, Kashef MA, Kashef S, Eghtedari F, Haffner K, Valmari P, Baktai G, Aviram M, Bentur L, Amirav I, Davis EE, Katsanis N, Brueckner M, Shaposhnykov A, Pigino G, Dworniczak B, Omran H. DNAH11 Localization in the Proximal Region of Respiratory Cilia Defines Distinct Outer Dynein Arm Complexes. American Journal of Respiratory Cell and Molecular Biology. 2016;55:213–224. doi: 10.1165/rcmb.2015-0353OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishikawa H, Thompson J, Yates JR, 3rd, Marshall WF. Proteomic analysis of mammalian primary cilia. Current biology : CB. 2012;22:414–419. doi: 10.1016/j.cub.2012.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayer U, Kuller A, Daiber PC, Neudorf I, Warnken U, Schnolzer M, Frings S, Mohrlen F. The proteome of rat olfactory sensory cilia. Proteomics. 2009;9:322–334. doi: 10.1002/pmic.200800149. [DOI] [PubMed] [Google Scholar]
- 23.Mick DU, Rodrigues RB, Leib RD, Adams CM, Chien AS, Gygi SP, Nachury MV. Proteomics of Primary Cilia by Proximity Labeling. Developmental cell. 2015;35:497–512. doi: 10.1016/j.devcel.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Narita K, Kozuka-Hata H, Nonami Y, Ao-Kondo H, Suzuki T, Nakamura H, Yamakawa K, Oyama M, Inoue T, Takeda S. Proteomic analysis of multiple primary cilia reveals a novel mode of ciliary development in mammals. Biol Open. 2012;1:815–825. doi: 10.1242/bio.20121081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pazour GJ, Agrin N, Leszyk J, Witman GB. Proteomic analysis of a eukaryotic cilium. J Cell Biol. 2005;170:103–113. doi: 10.1083/jcb.200504008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McClintock TS, Glasser CE, Bose SC, Bergman DA. Tissue expression patterns identify mouse cilia genes. Physiological genomics. 2008;32:198–206. doi: 10.1152/physiolgenomics.00128.2007. [DOI] [PubMed] [Google Scholar]
- 27.Li JB, Gerdes JM, Haycraft CJ, Fan Y, Teslovich TM, May-Simera H, Li H, Blacque OE, Li L, Leitch CC, Lewis RA, Green JS, Parfrey PS, Leroux MR, Davidson WS, Beales PL, Guay-Woodford LM, Yoder BK, Stormo GD, Katsanis N, Dutcher SK. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 2004;117:541–552. doi: 10.1016/s0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- 28.Geremek M, Bruinenberg M, Zietkiewicz E, Pogorzelski A, Witt M, Wijmenga C. Gene expression studies in cells from primary ciliary dyskinesia patients identify 208 potential ciliary genes. Human Genetics. 2011;129:283–293. doi: 10.1007/s00439-010-0922-4. [DOI] [PubMed] [Google Scholar]
- 29.Chapman JD, Goodlett DR, Masselon CD. Multiplexed and data-independent tandem mass spectrometry for global proteome profiling. Mass Spectrom Rev. 2014;33:452–470. doi: 10.1002/mas.21400. [DOI] [PubMed] [Google Scholar]
- 30.Panchaud A, Scherl A, Shaffer SA, von Haller PD, Kulasekara HD, Miller SI, Goodlett DR. Precursor acquisition independent from ion count: how to dive deeper into the proteomics ocean. Analytical chemistry. 2009;81:6481–6488. doi: 10.1021/ac900888s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gillet LC, Navarro P, Tate S, Rost H, Selevsek N, Reiter L, Bonner R, Aebersold R. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Molecular & cellular proteomics : MCP. 2012;11:O111 016717. doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silva JC, Denny R, Dorschel C, Gorenstein MV, Li GZ, Richardson K, Wall D, Geromanos SJ. Simultaneous qualitative and quantitative analysis of the Escherichia coli proteome: a sweet tale. Mol Cell Proteomics. 2006;5:589–607. doi: 10.1074/mcp.M500321-MCP200. [DOI] [PubMed] [Google Scholar]
- 33.Silva JC, Denny R, Dorschel CA, Gorenstein M, Kass IJ, Li GZ, McKenna T, Nold MJ, Richardson K, Young P, Geromanos S. Quantitative proteomic analysis by accurate mass retention time pairs. Anal Chem. 2005;77:2187–2200. doi: 10.1021/ac048455k. [DOI] [PubMed] [Google Scholar]
- 34.Silva JC, Gorenstein MV, Li GZ, Vissers JP, Geromanos SJ. Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol Cell Proteomics. 2006;5:144–156. doi: 10.1074/mcp.M500230-MCP200. [DOI] [PubMed] [Google Scholar]
- 35.Nahnsen S, Bielow C, Reinert K, Kohlbacher O. Tools for label-free peptide quantification. Molecular & cellular proteomics : MCP. 2013;12:549–556. doi: 10.1074/mcp.R112.025163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gevaert K, Impens F, Ghesquiere B, Van Damme P, Lambrechts A, Vandekerckhove J. Stable isotopic labeling in proteomics. Proteomics. 2008;8:4873–4885. doi: 10.1002/pmic.200800421. [DOI] [PubMed] [Google Scholar]
- 37.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nature biotechnology. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 38.Rauniyar N, Yates JR., 3rd Isobaric labeling-based relative quantification in shotgun proteomics. Journal of proteome research. 2014;13:5293–5309. doi: 10.1021/pr500880b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiese S, Reidegeld KA, Meyer HE, Warscheid B. Protein labeling by iTRAQ: a new tool for quantitative mass spectrometry in proteome research. Proteomics. 2007;7:340–350. doi: 10.1002/pmic.200600422. [DOI] [PubMed] [Google Scholar]
- 40.Shi T, Song E, Nie S, Rodland KD, Liu T, Qian WJ, Smith RD. Advances in targeted proteomics and applications to biomedical research. Proteomics. 2016;16:2160–2182. doi: 10.1002/pmic.201500449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kirkpatrick DS, Gerber SA, Gygi SP. The absolute quantification strategy: a general procedure for the quantification of proteins and post-translational modifications. Methods. 2005;35:265–273. doi: 10.1016/j.ymeth.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 42.Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Molecular & cellular proteomics : MCP. 2005;4:1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- 43.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 44.Fulcher ML, Gabriel S, Burns KA, Yankaskas JR, Randell SH. Well-differentiated human airway epithelial cell cultures. Methods Mol Med. 2005;107:183–206. doi: 10.1385/1-59259-861-7:183. [DOI] [PubMed] [Google Scholar]
- 45.Fulcher ML, Randell SH. Human nasal and tracheo-bronchial respiratory epithelial cell culture. Methods in molecular biology. 2013;945:109–121. doi: 10.1007/978-1-62703-125-7_8. [DOI] [PubMed] [Google Scholar]
- 46.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nature methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gavet O, Pines J. Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Developmental cell. 2010;18:533–543. doi: 10.1016/j.devcel.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCloy RA, Rogers S, Caldon CE, Lorca T, Castro A, Burgess A. Partial inhibition of Cdk1 in G 2 phase overrides the SAC and decouples mitotic events. Cell Cycle. 2014;13:1400–1412. doi: 10.4161/cc.28401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilm M, Shevchenko A, Houthaeve T, Breit S, Schweigerer L, Fotsis T, Mann M. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature. 1996;379:466–469. doi: 10.1038/379466a0. [DOI] [PubMed] [Google Scholar]
- 51.Cheng FY, Blackburn K, Lin YM, Goshe MB, Williamson JD. Absolute protein quantification by LC/MS(E) for global analysis of salicylic acid-induced plant protein secretion responses. Journal of proteome research. 2009;8:82–93. doi: 10.1021/pr800649s. [DOI] [PubMed] [Google Scholar]
- 52.Lee L, Campagna DR, Pinkus JL, Mulhern H, Wyatt TA, Sisson JH, Pavlik JA, Pinkus GS, Fleming MD. Primary ciliary dyskinesia in mice lacking the novel ciliary protein Pcdp1. Mol Cell Biol. 2008;28:949–957. doi: 10.1128/MCB.00354-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kultgen PL, Byrd SK, Ostrowski LE, Milgram SL. Characterization of an a-kinase anchoring protein in human ciliary axonemes. Mol Biol Cell. 2002;13:4156–4166. doi: 10.1091/mbc.E02-07-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nesvizhskii AI, Aebersold R. Interpretation of shotgun proteomic data: the protein inference problem. Molecular & cellular proteomics : MCP. 2005;4:1419–1440. doi: 10.1074/mcp.R500012-MCP200. [DOI] [PubMed] [Google Scholar]
- 55.Jensen-Smith HC, Luduena RF, Hallworth R. Requirement for the betaI and betaIV tubulin isotypes in mammalian cilia. Cell Motility and the Cytoskeleton. 2003;55:213–220. doi: 10.1002/cm.10122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vent J, Wyatt TA, Smith DD, Banerjee A, Luduena RF, Sisson JH, Hallworth R. Direct involvement of the isotype-specific C-terminus of beta tubulin in ciliary beating. Journal of cell science. 2005;118:4333–4341. doi: 10.1242/jcs.02550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pigino G, Bui KH, Maheshwari A, Lupetti P, Diener D, Ishikawa T. Cryoelectron tomography of radial spokes in cilia and flagella. The Journal of cell biology. 2011;195:673–687. doi: 10.1083/jcb.201106125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Urbanska P, Song K, Joachimiak E, Krzemien-Ojak L, Koprowski P, Hennessey T, Jerka-Dziadosz M, Fabczak H, Gaertig J, Nicastro D, Wloga D. The CSC proteins FAP61 and FAP251 build the basal substructures of radial spoke 3 in cilia. Molecular biology of the cell. 2015;26:1463–1475. doi: 10.1091/mbc.E14-11-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Knowles MR, Ostrowski LE, Leigh MW, Sears PR, Davis SD, Wolf WE, Hazucha MJ, Carson JL, Olivier KN, Sagel SD, Rosenfeld M, Ferkol TW, Dell SD, Milla CE, Randell SH, Yin W, Sannuti A, Metjian HM, Noone PG, Noone PJ, Olson CA, Patrone MV, Dang H, Lee HS, Hurd TW, Gee HY, Otto EA, Halbritter J, Kohl S, Kircher M, Krischer J, Bamshad MJ, Nickerson DA, Hildebrandt F, Shendure J, Zariwala MA. Mutations in RSPH1 cause primary ciliary dyskinesia with a unique clinical and ciliary phenotype. Am.J.Respir.Crit.Care Med. 2014;189:707–717. doi: 10.1164/rccm.201311-2047OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu F, Feng Y, Li Z, Pan C, Su Y, Yang R, Song L, Duan H, Deng N. Clinic-genomic association mining for colorectal cancer using publicly available datasets. Biomed Res Int. 2014;2014:170289. doi: 10.1155/2014/170289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sherlock G, Hernandez-Boussard T, Kasarskis A, Binkley G, Matese JC, Dwight SS, Kaloper M, Weng S, Jin H, Ball CA, Eisen MB, Spellman PT, Brown PO, Botstein D, Cherry JM. The Stanford Microarray Database. Nucleic Acids Research. 2001;29:152–155. doi: 10.1093/nar/29.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gupta M, Neavin D, Liu D, Biernacka J, Hall-Flavin D, Bobo WV, Frye MA, Skime M, Jenkins GD, Batzler A, Kalari K, Matson W, Bhasin SS, Zhu H, Mushiroda T, Nakamura Y, Kubo M, Wang L, Kaddurah-Daouk R, Weinshilboum RM. TSPAN5, ERICH3 and selective serotonin reuptake inhibitors in major depressive disorder: pharmacometabolomics-informed pharmacogenomics. Mol Psychiatry. 2016;21:1717–1725. doi: 10.1038/mp.2016.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lindskog C, Fagerberg L, Hallstrom B, Edlund K, Hellwig B, Rahnenfuhrer J, Kampf C, Uhlen M, Ponten F, Micke P. The lung-specific proteome defined by integration of transcriptomics and antibody-based profiling. Faseb J. 2014;28:5184–5196. doi: 10.1096/fj.14-254862. [DOI] [PubMed] [Google Scholar]
- 64.Delgado AP, Brandao P, Hamid S, Narayanan R. Mining the Dark Matter of the Cancer Proteome for Novel Biomarkers. Current Cancer Therapy Reviews. 2013;9:265–277. [Google Scholar]
- 65.Deane JA, Cole DG, Seeley ES, Diener DR, Rosenbaum JL. Localization of intraflagellar transport protein IFT52 identifies basal body transitional fibers as the docking site for IFT particles. Current biology : CB. 2001;11:1586–1590. doi: 10.1016/s0960-9822(01)00484-5. [DOI] [PubMed] [Google Scholar]
- 66.Rosenbaum JL, Witman GB. Intraflagellar transport. Nat Rev Mol Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- 67.Wang Z, Fan ZC, Williamson SM, Qin H. Intraflagellar transport (IFT) protein IFT25 is a phosphoprotein component of IFT complex B and physically interacts with IFT27 in Chlamydomonas. PLoS One. 2009;4:e5384. doi: 10.1371/journal.pone.0005384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Amira N, Cancel-Tassin G, Bernardini S, Cochand-Priollet B, Bittard H, Mangin P, Fournier G, Latil A, Cussenot O. Expression in bladder transitional cell carcinoma by real-time quantitative reverse transcription polymerase chain reaction array of 65 genes at the tumor suppressor locus 9q34.1–2: identification of 5 candidates tumor suppressor genes. Int J Cancer. 2004;111:539–542. doi: 10.1002/ijc.20283. [DOI] [PubMed] [Google Scholar]
- 69.Boelens MC, van den Berg A, Fehrmann RS, Geerlings M, de Jong WK, te Meerman GJ, Sietsma H, Timens W, Postma DS, Groen HJ. Current smoking-specific gene expression signature in normal bronchial epithelium is enhanced in squamous cell lung cancer. The Journal of pathology. 2009;218:182–191. doi: 10.1002/path.2520. [DOI] [PubMed] [Google Scholar]
- 70.Fong KW, Leung JW, Li Y, Wang W, Feng L, Ma W, Liu D, Songyang Z, Chen J. MTR120/KIAA1383, a novel microtubule-associated protein, promotes microtubule stability and ensures cytokinesis. Journal of cell science. 2013;126:825–837. doi: 10.1242/jcs.116137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bhattacharya R, Devi MS, Dhople VM, Jesudasan RA. A mouse protein that localizes to acrosome and sperm tail is regulated by Y-chromosome. BMC Cell Biol. 2013;14:50. doi: 10.1186/1471-2121-14-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Burkhard P, Stetefeld J, Strelkov SV. Coiled coils: a highly versatile protein folding motif. Trends in cell biology. 2001;11:82–88. doi: 10.1016/s0962-8924(00)01898-5. [DOI] [PubMed] [Google Scholar]
- 73.Dymek EE, Smith EF. A conserved CaM- and radial spoke associated complex mediates regulation of flagellar dynein activity. The Journal of cell biology. 2007;179:515–526. doi: 10.1083/jcb.200703107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lin J, Yin W, Smith MC, Song K, Leigh MW, Zariwala MA, Knowles MR, Ostrowski LE, Nicastro D. Cryo-electron tomography reveals ciliary defects underlying human RSPH1 primary ciliary dyskinesia. Nat Commun. 2014;5:5727. doi: 10.1038/ncomms6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Strand SH, Orntoft TF, Sorensen KD. Prognostic DNA methylation markers for prostate cancer. Int J Mol Sci. 2014;15:16544–16576. doi: 10.3390/ijms150916544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.P DA, Brandao P, Hamid S, Narayanan R. Mining the Dark Matter of the Cancer Proteome for Novel Biomarkers. Current Cancer Therapy Reviews. 2013;9:265–277. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.