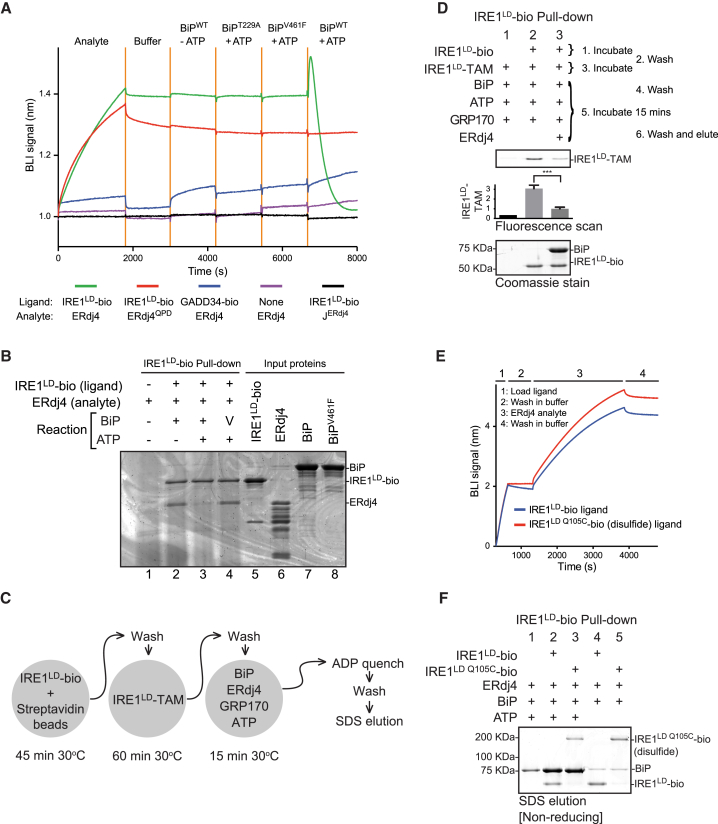
Figure 5.
ERdj4 Recruits BiP to Disrupt IRE1 Dimerization
(A) Bio-layer interferometry (BLI) signal of streptavidin sensors loaded with the indicated biotinylated ligand and reacted sequentially with the indicated solution of analyte, followed sequentially by the indicated solutions of BiP and ATP. Concentrations used were 1.5 μM ERdj4, 1 μM BiP, and 2 mM ATP.
(B) Protein recovered from a BLI sensor lacking (lane 1) or containing an IRE1LD-bio ligand (lanes 2–4). The sensor was incubated with an ERdj4 analyte and then with BiP or BiPV461F ± ATP.
(C) Schema of the experiment shown in (D).
(D) Fluorescence scans and Coomassie-stained SDS-PAGE gel of proteins recovered on immobilized streptavidin from reactions assembled from the indicated components. The IRE1LD-bio-loaded streptavidin beads were pre-associated with IRE1LD-TAM and then incubated in a solution of BiP, ERdj4, GRP170, and ATP. Bars show mean IRE1LD-TAM signal recovered with IRE1LD-bio (± SD) from four independent experiments, ∗∗∗p = 0.001 by parametric student’s paired ratio t test.
(E) Time-dependent changes in BLI signal of sensors loaded with either WT biotinylated IRE1LD (blue trace) or covalent dimeric disulfide-linked IRE1LD Q105C-bio (red trace) ligands. Ligand loading (step 1), wash (step 2), interaction with ERdj4 (step 3), and wash (step 4) are shown.
(F) Coomassie stained non-reducing SDS-PAGE gel of IRE1LD-bio and BiP recovered on a streptavidin matrix from reactions constituted as in Figures 4B and 4C but with IRE1LD-bio or covalent dimeric disulfide-linked IRE1LD Q105C-bio. Proteins were eluted with SDS sample buffer.
See also Figure S4.