Abstract
Dicer-dependent small noncoding RNAs play important roles in gene regulation in a wide variety of organisms. Endogenous small interfering RNAs (siRNAs) are part of an ancient pathway of transposon control in plants and animals. The ciliate, Oxytricha trifallax, has approximately 16,000 gene-sized chromosomes in its somatic nucleus. Long noncoding RNAs establish high ploidy levels at the onset of sexual development, but the factors that regulate chromosome copy numbers during cell division and growth have been a mystery. We report a novel function of a class of Dicer (Dcl-1)- and RNA-dependent RNA polymerase (RdRP)-dependent endogenous small RNAs in regulating chromosome copy number and gene dosage in O. trifallax. Asexually growing populations express an abundant class of 21-nt sRNAs that map to both coding and noncoding regions of most chromosomes. These sRNAs are bound to chromatin and their levels surprisingly do not correlate with mRNA levels. Instead, the levels of these small RNAs correlate with genomic DNA copy number. Reduced sRNA levels in dcl-1 or rdrp mutants lead to concomitant reduction in chromosome copy number. Furthermore, these cells show no signs of transposon activation, but instead display irregular nuclear architecture and signs of replication stress. In conclusion, Oxytricha Dcl-1 and RdRP-dependent small RNAs that derive from the somatic nucleus contribute to the maintenance of gene dosage, possibly via a role in DNA replication, offering a novel role for these small RNAs in eukaryotes.
Keywords: DNA copy number, Dicer, epigenetics, small RNAs, ciliates
INTRODUCTION
Small RNAs that associate with the conserved PIWI family of Argonaute proteins are important regulators of gene expression in a broad range of RNA silencing pathways (Ghildiyal and Zamore 2009). The sequence information in small noncoding RNAs provides the specificity for homology-dependent gene silencing. One such class of small RNAs, small interfering RNAs (siRNAs), typically 19- to 25-nt long, derive from cleavage of long double-stranded RNAs (dsRNAs) by Dicer, an RNase III type enzyme (Bernstein et al. 2001). Together, siRNAs bound to Argonaute proteins form an RNA-induced silencing complex (RISC) (Hammond et al. 2000), responsible for silencing target mRNAs in the process known as RNA interference (RNAi) (Fire et al. 1998).
In many organisms, RNA-dependent RNA polymerases (RdRP) amplify the Dicer-generated “primary” siRNAs to produce “secondary” siRNAs that further enhance the silencing effect (Dalmay et al. 2000; Sijen et al. 2001; Volpe et al. 2002). While dsRNA viruses often trigger RNAi, endogenous siRNAs can target developmentally regulated genes, as well as pseudogenes, and are also crucial for transposon silencing (Czech et al. 2008; Ghildiyal et al. 2008; Kawamura et al. 2008; Okamura et al. 2008; Tam et al. 2008; Axtell 2013). In yeast and plants, they are additionally involved in heterochromatin formation and transgene silencing (Castel and Martienssen 2013). The ciliate Tetrahymena expresses diverse small RNA classes that are derived from either pseudogene clusters, protein coding genes, or telomeres (Couvillion et al. 2009; Farley and Collins 2017). Both Tetrahymena and Paramecium express a meiosis-specific, Dicer-dependent class of small RNAs called scanRNAs that derive from the germline micronucleus (Mochizuki et al. 2002; Lepère et al. 2009). Irrespective of their biological function, the endogenous siRNAs usually act at the transcriptional or post-transcriptional level to silence their target genes.
Oxytricha trifallax, a spirotrich ciliate, has dimorphic nuclei within the same cell (Prescott 1994). The macronucleus (MAC) contains the somatic genome, responsible for gene transcription during asexual growth, while the micronucleus (MIC) is the zygotic nucleus, which supplies both an archival germline nucleus and a duplicate copy that develops into the new macronucleus. An arsenal of noncoding RNAs, ranging from long telomere-containing transcripts (Nowacki et al. 2008; Lindblad et al. 2017) to 27-nt piRNAs (Fang et al. 2012) regulates the whole process of genome rearrangement to produce a new somatic genome during post-zygotic development.
The macronuclear genome harbors approximately 16,000 gene-sized chromosomes, most of which contain only a single gene and are present at a very high copy number (Swart et al. 2013). The MAC is also remarkably devoid of repetitive elements like transposons, which are replete in the micronucleus (Chen et al. 2014). Lacking in spindle microtubules (Fujiu and Numata 2000), the macronucleus divides amitotically during asexual cell divisions, with no known mechanism to segregate its somatic chromosomes to daughter cells. Here, we report a novel role for endogenous sRNAs in asexually growing Oxytricha cells in the maintenance of DNA dosage and chromosome copy number. Oxytricha sRNA sequences derive from both coding and noncoding regions for almost all fully sequenced somatic chromosomes. Disruption of a Dicer-like gene, dcl-1, leads to reduced levels of sRNAs; however, the resulting cells show no sign of post-transcriptional gene silencing (PTGS) or transposon reactivation. Instead, they display reduced DNA copy number and signs of S-phase arrest and DNA breakage. These observations reveal a novel role for Dicer-dependent sRNAs in gene dosage and maintenance of chromosome copy number in Oxytricha.
RESULTS
Dcl-1 and RdRP-dependent small RNAs in asexually growing Oxytricha cells
The Oxytricha macronuclear genome encodes one Dicer and three Dicer-like (Dcl) genes, all of which are expressed at varying levels during asexual, vegetative growth (Supplemental Fig. S1A,B; Swart et al. 2013). Four RNA-dependent RNA polymerase (RdRP) genes are also encoded in the MAC genome. Two of them, encoded on Contig14356.0 and Contig17273.0, are expressed at very low levels and could be pseudogenes (Supplemental Fig. S1C). Contig20928.0 and Contig4554.0 are expressed at much higher levels and likely function during development (Supplemental Fig. S1C). Using a synthetic piRNA injection strategy (based on Fang et al. 2012 and described in Materials and Methods), we prepared epigenetically altered strains expressing somatic mutant versions of Dcl and RdRP. This allowed us to query their roles in the biogenesis and function of these small RNAs. Disruption of dcl-2 (Contig119.0), the most highly expressed Dicer-like gene, failed to produce any viable cells, possibly due to essential functions of the protein during conjugation or vegetative growth. Alterations to dcl-1, the second most highly expressed Dicer-like gene in Oxytricha, yielded some clonal lines with complete disruption of the open reading frame via programmed retention of a normally deleted (germline-limited) 24-bp sequence between the 622nd and 623rd codon of the gene (Supplemental Fig. S2A,B). The resulting insertion creates a premature termination codon, which would produce a truncated protein lacking the dsRNA binding and Ribonuclease III (RNase III) domains (Supplemental Fig. S2C). Similarly, we introduced functional mutations into the somatic RdRP gene using sRNA injection to program a frameshift, and analyzed mutants with complete reading frame disruption (Supplemental Fig. S2D–F).
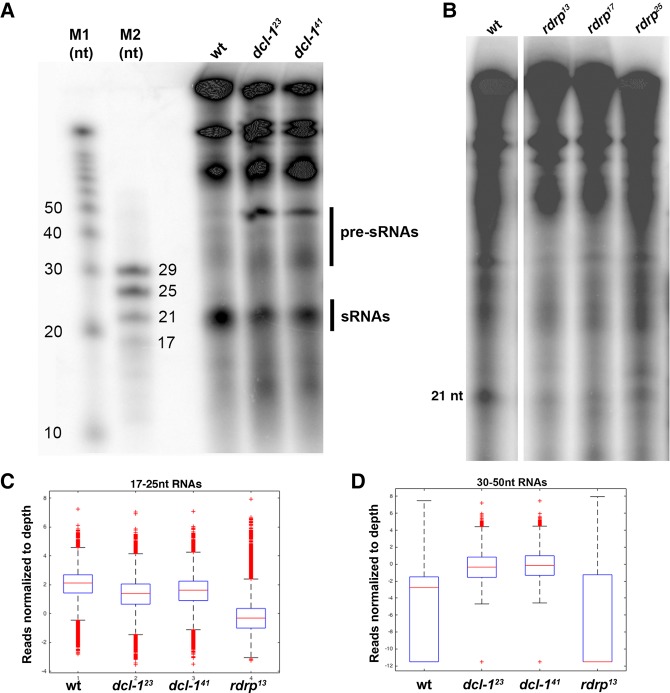
To assess their function in small RNA biogenesis, total RNA from wild-type strain JRB310 as a control, two dcl-1 mutant strains (dcl-123 and dcl-141) and three rdrp strains (rdrp13, rdrp17, and rdrp25) was radiolabeled with [γ-32P]-ATP and separated on a denaturing polyacrylamide gel, followed by phosphorimaging analysis. The dcl-1 cells showed reduced levels of 21-nt small RNAs and increased accumulation of 30- to 50-nt RNA (Fig. 1A). rdrp mutant lines also displayed reduced 21-nt RNA levels relative to wild-type control cells (Fig. 1B), demonstrating that sRNA production in asexually growing Oxytricha cells is dependent on both Dcl-1 and RdRP. The sRNA reduction is partial, suggesting that other paralogous Dicer or RdRP genes might provide redundant functions with the mutants investigated in this study.
FIGURE 1.
Dcl-1 and RdRP-dependent small RNAs in vegetatively growing Oxytricha cells. (A) Twenty-one nucleotide sRNAs are dependent on Dcl-1 and (B) RdRP genes. In addition, 30- to 50-nt RNAs accumulate in dcl-1 strains. (C) Box plot displaying reduced levels of MAC mapping 17- to 25-nt sRNAs in dcl-123, dcl-141, and rdrp13 cells versus wild type (JRB310), shown as a reduction in sRNA reads mapping to MAC contigs, whereas (D) 30- to 50-nt putative sRNA precursors accumulate in dcl-123 and dcl-141. Coverage calculated for 12,203 fully assembled MAC chromosomes and normalized based on total sequencing depth of all samples. The Wilcoxon rank sum test was used to confirm statistical significance. P-values for 17- to 25-nt sRNAs for all pairwise comparisons between WT and mutants are <10−5.
Notably, rdrp mutants do not display an increased accumulation of 30- to 50-nt RNAs, which is consistent with RdRP's activity in generating precursor substrates for small RNAs. This suggests that the small RNAs are Dcl-1- and RdRP-dependent, while the 30- to 50-nt RNA could contain putative sRNA precursors.
Total RNA from both wild-type, dcl-1, and rdrp mutants was isolated and small RNAs sequenced on an Illumina platform. Following read preprocessing (see Materials and Methods), the RNA sequences were aligned to the Oxytricha macronuclear (Swart et al. 2013), micronuclear (Chen et al. 2014), and mitochondrial (Swart et al. 2012) genome assemblies using Bowtie2 (Langmead and Salzberg 2012). Un-aligned sequences were mapped to a bacterial genome database downloaded from NCBI and the Chlamydomonas reinhardtii genome assembly (Merchant et al. 2007) to filter out environmental or food contaminants, respectively. The remaining unmapped sequences were classified as “Other.” Comparison of 17- to 25-nt RNAs from wild-type versus mutant cells revealed reduced proportions of 21-nt RNAs mapping to 10,513 fully sequenced MAC chromosomes in dcl-141 cells (Fig. 1C). However, the sRNA mapping and sequence characteristics remain unaltered in dcl-1 or rdrp mutant cells (Supplemental Fig. S3A–D); only their relative abundance is affected, compared to wild type. Together, our observations suggest that this novel class of small RNAs in Oxytricha may be homologous to siRNAs in other eukaryotes.
Oxytricha 21-nt sRNAs do not regulate mRNA levels
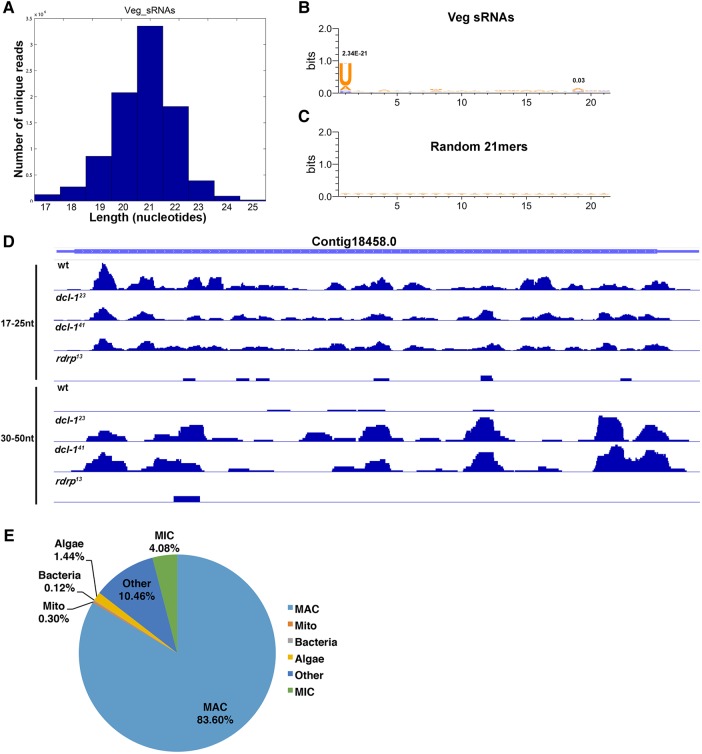
The small RNA length distribution in both WT and mutant cells reveals a peak at 21 nt and a statistically significant presence of U at the first position, both characteristics of siRNAs in other organisms (Fig. 2A–C). Small RNAs from asexually growing “vegetative” cells (Veg) map primarily to the MAC genome. Mapping of the sRNAs to the set of fully assembled two-telomere MAC chromosomes revealed an average of 2× genome coverage (Swart et al. 2013). Small RNAs map to both DNA strands with a bias toward the 5′ end of the transcription start site (TSS) on single-gene chromosomes (Supplemental Fig. S4A). Similarly, sRNA mapping to two-gene chromosomes shows a bias toward the TSS (Supplemental Fig. S4B–E). No small RNAs map to the telomere repeats. Figure 2D shows the mapping of sRNAs from wild-type, dcl-1 (dcl-123 and dcl-141), and rdrp (rdrp13) cells along a representative MAC Contig18458.0, which displays features typical of most MAC genes and encodes a glycosyltransferase family protein. Both dcl-1 and rdrp mutants show reduced 17- to 25-nt sRNAs versus wild-type control cells, while dcl-1 cells display an overabundance of 30- to 50-nt RNAs, which are undetectable in wild-type or rdrp mutants (Figs. 1D, 2D). Genome-wide, small RNAs map to both introns and sub-telomeric regions, as well as to coding regions (Supplemental Fig. S3E,F).
FIGURE 2.
Oxytricha MAC sRNAs do not lead to post-transcriptional gene silencing. (A) The length distribution profile for sRNAs shows a peak at 21 nt. Sequence logos for (B) 21-nt sRNAs versus (C) random 21-nt words generated from the macronuclear sequence show a strong preference for U at the first position (P-value = 2.3 × 10−21) and a modest increase in A at the 19th position (P-value = 0.03); P-values noted above the 1st and 19th bases. (D) 17- to 25-nt and 30- to 50-nt sRNA mapping to representative MAC Contig18458.0 for wt, dcl-1 and rdrp cells show reads mapping to the entire chromosome (excluding telomeres), including the CDS (thickest bars) and nongenic, subtelomeric regions (thin, flanking lines). (E) sRNA mapping statistics reveal that the majority of sRNAs map to the MAC genome (MIC, germline-limited; Mito, mitochondria). Of the fraction of sRNAs that map to germline-limited regions, 0.5% map to IESs and 1.6% to transposons or other MIC-limited repeats. Sequences in the “Other” category do not map to any Oxytricha genome.
The sRNA sequences preferentially map to the MAC genome (83.6%). 4.28% map to rearrangement junctions that are only present in the MAC. To exclude sequence-specific biases, a random set of MAC-derived 5′-U containing 21-nt sequences was generated and mapped to the genome. 3.87% of these randomized sequences map to the MAC-specific rearrangement junctions, which is not significantly different from the 4.28% of sRNA reads that map to these locations. A negligible fraction of sRNA reads map to the mitochondria (0.3%). 4.1% map to the MIC-limited (i.e., deleted) portion of the MIC genome (Fig. 2E; Supplemental Fig. S5) and will be discussed below. Note that 0.31% map to MIC-specific junction reads, whereas 1.25% of randomly sampled 21-nt 5′-U sequences in the MIC map to MIC-specific junctions. Therefore, most sRNA reads derive from the MAC, which suggests a potential regulatory role for these small RNAs in somatic gene expression (further details of MAC versus MIC mapping are provided in Materials and Methods).
Since dcl-1 and rdrp mutants displayed reduced levels of MAC-mapping sRNAs, we examined these sRNAs in greater detail. A genome-wide analysis (n = 12,203 completely sequenced MAC chromosomes) revealed only a modest bias for sRNAs derived from coding regions (Supplemental Fig. S3E) relative to the MAC genome-wide average (Supplemental Fig. S3F). These observations suggest that, rather than deriving from mRNA precursors, these sRNAs, like Oxytricha piRNAs (Fang et al. 2012), could be processed from long, noncoding RNAs, possibly from the same RNA molecules that establish chromosome copy number post-conjugation (Nowacki et al. 2010).
Analysis of the 30- to 50-nt RNAs that accumulate in dcl-1 cells revealed that they share similar properties with the mature 21-nt sRNAs, including mapping to entire MAC contigs (56.1%) and comparable coverage of coding versus noncoding regions. To confirm their genomic source, we compared the fraction of 30- to 50-nt reads mapping to MAC- and MIC-specific regions. 16,580 MAC-mapping reads (4.5%) derive from rearrangement junctions, which are MAC-specific. Note that 8.5% of a randomized sample of 40-nt words from two-telomere containing MAC sequences map to such junctions; however, a slight bias among the reads for transcription start sites may contribute to this difference. Just 0.3% of MIC-mapping reads map to MIC-limited rearrangement junctions, whereas 2.6% of randomly generated 40-nt MIC sequences map to MIC-limited junctions. Together, these and the above analyses suggest that the sRNAs derive from the MAC and that the 30- to 50-nt RNAs may be MAC-derived sRNA precursors, like pre-siRNAs.
Additional evidence that the Oxytricha sRNAs do not lead to PTGS, comes from the observation that sRNA levels do not correlate with RNA-seq levels from similar cell populations (Pearson correlation coefficient, R = 0.11) (Supplemental Fig. S6; Swart et al. 2013). Furthermore, to test for global changes in gene expression, we performed RNA sequencing (RNA-seq) from the poly(A)-enriched fraction of total RNA from two biological replicates for WT, dcl-141, and rdrp13 cells. Differential expression analysis (DE) using edgeR revealed only a handful of genes affected (discussed below) (Supplemental File 1). This is contrary to expectations if a role of the sRNAs were to down-regulate mRNA levels. This suggests that the sRNAs do not inhibit somatic gene expression, but instead might directly or indirectly activate mRNA expression.
Exogenous addition of long double-stranded RNA (dsRNA) in Oxytricha and other ciliates can lead to mRNA repression in a sequence-dependent manner, via RNA interference (RNAi). This is typically performed by feeding Oxytricha cells E. coli expressing the target gene within a dsRNA producing plasmid. cDNA regions of three target genes, Cbx5, Actin, and Ku80, were cloned into a dsRNA producing plasmid vector, L4440, and transformed into the RNase III-deficient E. coli strain HT115. Upon RNA induction, heat-killed bacteria were fed to Oxytricha cells using established RNAi protocols for conjugating cells (Nowacki et al. 2008). Treated Oxytricha cells were exclusively fed for 2 d on induced E. coli, and total RNA and DNA were isolated the next day. Surprisingly, none of these dsRNA feeding experiments led to even twofold reduction in mRNA levels for the target genes (Supplemental Fig. S7A). Taken together, the above results suggest that the 21-nt Oxytricha sRNAs produced in asexually growing cells are not involved in mRNA silencing.
RdRP-dependent MIC-derived small RNAs
The minor fraction of MIC-specific small RNAs (Supplemental Fig. S5A,B) have a length distribution peak at 22 nt, in contrast to the 21-nt peak for sRNAs that map to the MAC. Approximately 47% of the MIC genome is repetitive DNA (Chen et al. 2014). Telomere-bearing elements (TBEs) are the most abundant class of DNA transposons in the Oxytricha genome, representing ∼15% of the MIC genome, and 30% of the MIC-mapping sRNA reads map to TBE transposons. However, these small RNAs lack the characteristic peak at 21–22 nt, suggesting they could be degraded RNA. In contrast, small RNAs that map to the most abundant classes of satellite repeats show a distinct peak at 22 nt (Supplemental Fig. S5A). Approximately 1% of the MIC genome encodes single-copy, germline-limited genes (Chen et al. 2014), but 5% of MIC-mapping sRNAs map to these genic loci (Supplemental Fig. S5B), suggesting that these small RNAs could be involved in silencing germline gene expression in asexually growing cells. In contrast to MAC-mapping sRNAs, dcl-1 cells show no reduction in the levels of MIC-mapping sRNAs (Supplemental Fig. S5C,D). However, rdrp cells showed a small yet significant reduction in MIC-satellite and MIC gene mapping small RNAs (Supplemental Fig. S5C,D). To test whether these changes could affect gene expression, we mapped our RNA sequencing (RNA-seq) data to various MIC genomic loci. Differential expression analysis (DE) using edgeR showed increased expression of a subset of MIC satellites in rdrp13 compared to control (Supplemental Fig. S5E). In contrast, dcl-141 cells showed no change in MIC-satellite expression level, consistent with no change in MIC-derived small RNAs in those samples. No significant MIC gene expression was observed in either of the samples, suggesting that either our coverage was not enough to detect low transcript levels or that those genes are likely silenced by other redundant mechanisms in rdrp13 cells. Genomic DNA sequencing of both wild-type and dcl-1 cells also showed no evidence of increased transposon activity (0.1% of reads map to TBEs in both WT and dcl-1). Taken together, these findings reveal genetically distinct classes of small RNAs in asexually growing Oxytricha cells; Dcl-1-dependent MAC-mapping 21-nt sRNAs and RdRP-dependent MIC-mapping 21- to 22-nt sRNAs.
Oxytricha 21-nt sRNAs regulate DNA copy number
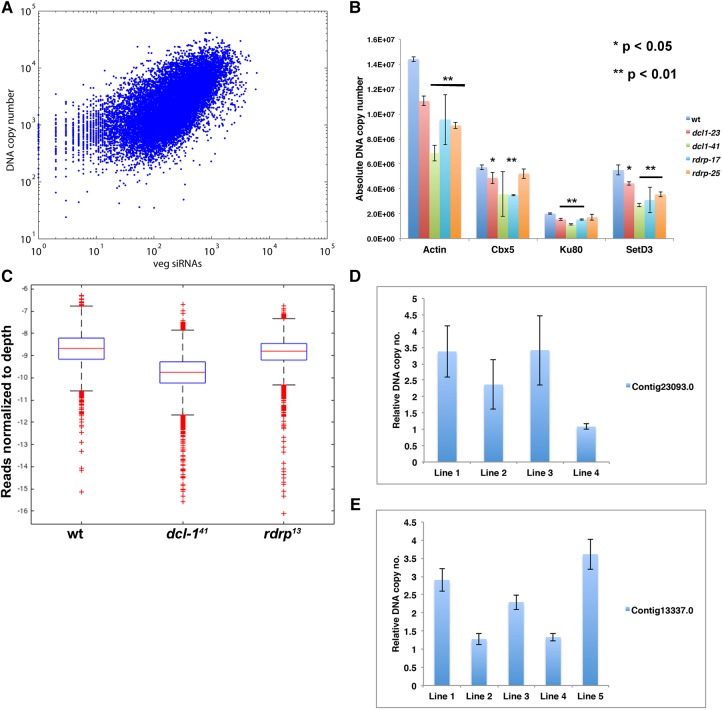
Notably, instead, we observed a positive correlation between sRNA levels and DNA copy number (Pearson correlation coefficient, R = 0.662), which suggests that the presence of sRNAs could positively regulate DNA copy number (Fig. 3A). Though dsRNA feeding does not lead to mRNA repression, we observed an increase in DNA copy number in cells fed bacteria expressing Oxytricha-specific dsRNAs, relative to an empty vector control, suggesting that the increased availability of sRNAs for target genes could potentially be a positive regulator of gene dosage (Supplemental Fig. S7B). This effect of increased copy number was dependent on Dcl-1. We chose four different MAC contigs to represent the range of low and high DNA copy-number levels. Absolute DNA quantification was performed using qPCR and the data were normalized to the levels of mitochondrial rDNA, whose levels are relatively constant between different wild-type and mutant strains (Supplemental Fig. S7C). Quantitative analysis of DNA copy number in control versus mutant strains revealed statistically significant DNA copy number reductions for these different MAC loci in one or both mutant alleles of dcl-1 and rdrp, suggesting the presence of an Oxytricha siRNA-like pathway involved in DNA copy-number regulation (Fig. 3B).
FIGURE 3.
Oxytricha 21-nt MAC sRNAs correlate with genomic DNA copy number. (A) MAC DNA copy number and sRNA copy number in asexually growing cells (veg) display a positive correlation (R = 0.662). (B) qPCR analysis of absolute DNA quantification for four representative MAC contigs shows statistically significant reduction of DNA copy number in dcl-1 and rdrp mutant strains. Error bars represent standard deviation from the mean for three experiments. Equal amounts of DNA were taken for each of the samples. (*) P-value <0.05; (**) P-value <0.01. (C) Box plot displaying DNA copy number between wt, dcl-141, and rdrp13 (all exconjugant cells) shows a genome-wide reduction in MAC DNA copy number in dcl-1 and rdrp cells. Copy number is based on mapped reads normalized to depth, further normalized by mitochondrial rDNA reads to account for differences in amount of DNA. P-values for pairwise comparisons yield values either less than 10−5 or are below the detection limit. DNA copy-number analysis of cells injected with sRNAs against (D) Contig 23093.0 or (E) Contig13337.0, normalized against GFP control sRNA injected cells, demonstrates a 2.4-fold (D) or 2.28-fold (E) average increase in DNA levels in injected cells across all biological replicates. Four (Contig 23093.0) or five (Contig13337.0) different cell lines were established from single injected cells. Lines that demonstrate no change could have been a failed injection. The data from each established line were averaged across three technical replicates.
We confirmed these results genome-wide by sequencing genomic DNA libraries prepared from wild-type, dcl-141 and rdrp13 mutant cells and comparing DNA copy number for all fully assembled MAC chromosomes in the JRB310 and JRB510 reference genomes (Swart et al. 2013; Chen et al. 2015). Out of 12,203 contigs assayed, 12,132 and 9331 contigs show reduced DNA copy number in dcl-1 and rdrp, respectively (Fig. 3C).
To confirm that the sRNAs can directly affect gene dosage, we microinjected synthetic 21-nt dsRNAs against two different chromosomes. These targets (Contig 13337.0 and Contig 23093.0) were chosen because of their small length and low inherent DNA copy number in wild-type cells. We targeted three duplexes roughly equidistant from each other. These duplexes were chosen to match endogenous sRNA characteristics, such as the presence of a 5′-uridine and similar AT content. Microinjection of synthetic, duplexed 21-nt sRNAs against Contig23093.0 led to a two- to 3.5-fold increase in its DNA copy levels in three out of four lines injected, relative to an unrelated dsRNA injection (Fig. 3D). Additionally, we confirmed that dsRNA injection does not affect the copy number of unrelated genes, suggesting that the effect of dsRNAs is specific to the targeted gene (Supplemental Fig. S7D).
Similarly, injection of sRNAs against Contig13337.0, which encodes a Zinc-finger protein, led to 2.5- to 3.5-fold increases in DNA dosage in three out of five injected lines, relative to negative control GFP RNA oligonucleotide injection (Fig. 3E). DNA copy-number regulation appears tightly controlled, as displayed by relatively similar MAC chromosome levels in different Oxytricha strains (Supplemental Fig. S7E). Thus, these DNA copy number increases upon dsRNA injection suggest that the Dcl-1- and RdRP-dependent 21-nt sRNAs have the potential to positively regulate DNA copy number in Oxytricha.
Oxytricha 21-nt sRNAs are associated with chromatin and replication
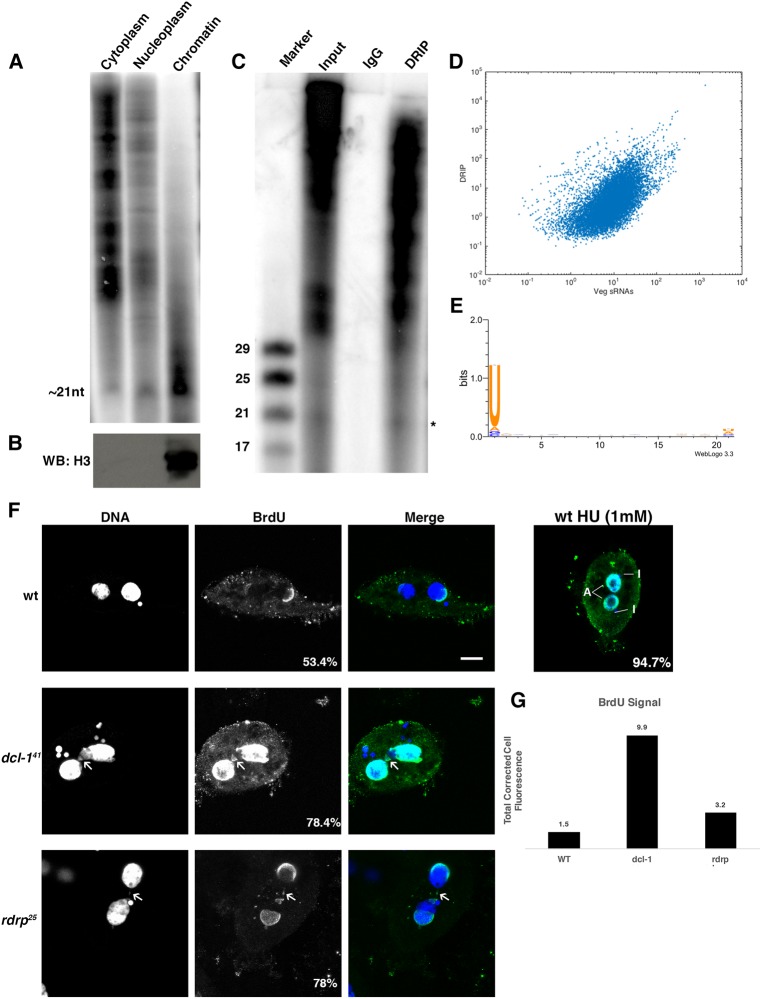
Mutant strains of dcl-1 and rdrp display slower growth and increased death rate with age (Supplemental Fig. S8). To help elucidate the mechanism of action for the 21-nt sRNAs, we examined their cellular localization. Cells were fractionated into cytoplasmic, nucleoplasmic and chromatin fractions and total RNA was isolated and radiolabeled. The 21-nt sRNAs are enriched in the chromatin fraction, suggesting a direct interaction with DNA (Fig. 4A). Proteins were also isolated from these fractions and immuno-hybridized to Histone H3, to confirm the purity of the fractions (Fig. 4B). These findings were further corroborated with a different methodology, via pull-down of DNA:RNA hybrids using a well-characterized S9.6 antibody (Fig. 4C; Phillips et al. 2013). For this, DNA:RNA hybrids were immunoprecipitated from formaldehyde-fixed chromatin extracts and RNA purified. Small RNA sequencing from immunoprecipitated DNA–RNA hybrids was performed and analyzed as described previously. The read counts mapping to two-telomere contigs correlate well between DRIP and veg sRNAs (R = 0.58) (Fig. 4D) and the 21-nt sRNAs display a similar 5′-U bias as total 21-nt sRNAs (Fig. 4E). This approach confirmed that the sRNAs associate with chromatin, where they might regulate or sense DNA copy number.
FIGURE 4.
Oxytricha 21-nt sRNAs are associated with chromatin and mutants display symptoms of S-phase arrest. (A) Purified RNA from cellular fractionation separated on a 15% denaturing polyacrylamide gel reveals chromatin enrichment of the 21-nt RNAs. (B) Western analysis (WB:H3) confirms the purity of the chromatin fraction by immunohybridization against Histone H3. (C) DNA.RNA hybrid immunoprecipitation (DRIP) from vegetative chromatin extracts also suggests that the 21-nt sRNAs (marked with [*]) associate with DNA. (D) Read counts for mapped vegetative (Veg) sRNAs correlate with those recovered from DRIP (R = 0.582). (E) Staining of indicated cell types with anti-BrdU (green in merge) and DAPi (blue in merge) for DNA shows S-phase arrest. Also, note that nuclear fragments (marked with white arrows) in sRNA mutants are BrdU positive. (F) 94.7% of wild-type cells treated for 24 h with 1 mM hydroxyurea (HU) and labeled for 5 h with BrdU stain positively for BrdU. MAC (A) and MIC (I) are labeled with a white line. (G) Signal across the BrdU channel was quantified using Fiji (https://imagej.net) and total cell corrected fluorescence was plotted for indicated samples.
To study potential cell cycle defects in the dcl-1 and rdrp mutant strains, we labeled newly replicated DNA using BromodeoxyUridine (BrdU) by adding BrdU directly in the cell culture for 5 h, followed by cell fixation, DNA denaturation and staining using an anti-BrdU antibody. Note that 48.9% of wild-type cells (40%–58.6% across four biological replicates) stain positively for BrdU, while 71.1% of dcl-1 cells (56.7%–86.6% over four replicates) and 78% of rdrp cells (70%–80% across three biological replicates) stain positively, with brighter staining intensity (Fig. 4E). This observation is similar to wild-type cells treated with the replication stress inducer hydroxyurea (HU), where 94.7% of HU treated cells stain positively for BrdU (Fig. 4F). The BrdU signal was quantified for all samples and represented as total corrected cell fluorescence (Fig. 4G). These observations hint that dcl-1 and rdrp mutants might display S-phase arrest. Additionally, we note that dcl-1 and rdrp mutants show increased numbers of BrdU-positive nuclear fragments, analogous to “micronuclei” seen in mammalian cells with compromised genome integrity (white arrow; Fig. 4D) (Terradas et al. 2010). In contrast, BrdU labeling in dcl-1 cells without DNA denaturation failed to label nuclear fragments (Supplemental Fig. S9). This observation suggests that sRNA mutants display compromised genome integrity and defects in cell-cycle progression.
To examine potential gene expression changes that might accompany such nuclear defects, we mapped RNA-seq data for mutant strains to two-telomere contigs and compared the different samples. Even though DNA damage response involves post-translational modifications on existing proteins, such as phosphorylation of Histone H2Ax and ATM kinase, we identified eight differentially expressed genes in dcl-1 versus WT, three of which are associated with DNA damage response (Arsenite pump driving ATPase Contig9927.0 and Contig15729.0; superoxide dismutase; Contig15813.0) (Supplemental Fig. S10; Supplemental File 1). The five other genes encode unknown proteins, with no clear homolog outside the Oxytricha lineage. Similarly, comparison with rdrp identified 90 DE genes. The only significant GO term associated with these genes was “sequence-specific DNA binding”. This list included all the genes identified in dcl-1 cells along with all the major histone genes (Supplemental Fig. S9; Supplemental File 1). Histone gene expression is known to be restricted to S phase of the cell cycle. An over-expression of histone genes is therefore consistent with the suggestion that the mutant cells arrest in S-phase. Taken together, our observations point to a novel function of 21-nt small RNAs in regulating gene dosage and maintenance of chromosome ploidy in Oxytricha.
DISCUSSION
Small noncoding RNAs bound to Argonaute proteins constitute an ancient and well-studied pathway for regulating gene expression in plants and animals, where the endogenous siRNA pathway is typically involved in silencing foreign genetic invaders, like viruses and transposons. Abrogation of components of this pathway usually leads to increased transposon activity and hence compromised genome integrity. Oxytricha’s dual genome architecture offers a different model system to study small RNA function. Here, we report a novel sRNA pathway involved in genome maintenance in Oxytricha during asexual growth. Mutations that stall the processing or production of 21-nt sRNAs do not lead to increased transposon activity or increased target mRNA levels, but instead lead to reduced DNA copy number, potential S-phase arrest, and DNA damage. Furthermore, the 21-nt sRNAs map to the entire somatic genome and associate with chromatin. Together, these observations suggest a possible involvement of Oxytricha’s 21-nt sRNAs in DNA replication. Alternatively, the sRNAs could act as DNA dosage sensors that facilitate segregation of somatic chromosomes to daughter cells following asexual cell division.
During the sexual phase of the life cycle, genome-wide transcription of the somatic chromosomes from the parental macronucleus produces long noncoding template RNAs that guide DNA rearrangements (Nowacki et al. 2008; Khurana et al. 2014; Lindblad et al. 2017) and also regulate chromosome copy number in the progeny (Nowacki et al. 2010); however, the mechanism is unclear. The new class of sRNAs reported in this manuscript could derive from these maternal template RNAs that appear post fertilization. Like siRNAs, these 21-nt sRNAs could then be stably maintained and amplified by RdRP to permit regulation of chromosome ploidy levels in asexually dividing cells, which is consistent with the observed defects in rdrp mutant cells. Further evidence supporting this model comes from the observation that small RNAs sequenced from developmental time points post-fertilization show a positive correlation (R = 0.61) between sRNA levels in late conjugation and in vegetative cells (JS Khurana and LF Landweber, unpubl.).
We note that the overall effect of sRNA pathway mutations on DNA copy number is relatively small. This could result from one of two possibilities. Either the cells with the strongest phenotype are depleted from the asexually growing population, consistent with the increased death rate among the mutants, or alternatively, the paralogs of genes under study may act redundantly in the DNA dosage pathway, thus ameliorating the effects we report here. On the other hand, the conjugation-specific lncRNAs (Nowacki et al. 2010) are potential substrates for these sRNAs, and their amplification could lead to a stronger downstream effect on the target DNA copy number.
Parallel observations arise in other systems. For example, in Drosophila, dcr-2 mutants show destabilization of the ribosomal DNA locus in the form of extrachromosomal circular DNA (eccDNA), which often accumulates due to DNA replication defects (Peng and Karpen 2007). In Neurospora, DNA damage induces expression of Argonaute QDE-2 interacting small RNAs (qiRNAs) and an increase in ribosomal RNA levels (Lee et al. 2009); however, it is unknown whether the ribosomal DNA (rDNA) copy number is influenced as well. More recently, studies in S. pombe reported that Dcr-1 plays an important role in maintenance of genome integrity by inhibiting DNA recombination at rDNA repeats (Castel et al. 2014). Furthermore, Dicer in S. pombe maintains genomic integrity at rDNA loci by promoting transcription termination (Castel et al. 2014). Oxytricha dcl-1 mutants show 3.3-fold reduced levels in small RNAs that map to the rDNA chromosome and a twofold reduction in rDNA copy number. Furthermore, because most genome surveys discard or deplete ribosomal RNA sequences, either during library preparation or in data analysis, it is possible that siRNAs, more generally, could play a conserved role in regulating the dosage of high copy number loci in other organisms, as well.
MATERIALS AND METHODS
Cell culture and mutant strain construction
Wild-type O. trifallax strains JRB310 and JRB510 were grown as described (Khurana et al. 2014). For mutant strain construction, 27-nt artificial piRNAs were microinjected into conjugating cells, 12–15 h post mixing, as described in Fang et al. (2012). Exposure to these synthetic RNAs that match short DNA sequences within a gene leads to the retention of those sequences in the progeny, also known as “IES+ strains” that we describe in more detail below.
The MAC genome is present within the MIC genome as hundreds of thousands of retained DNA fragments, called the MAC-destined sequences (MDS). These MDSs within the MIC are interrupted by intragenic regions called internally eliminated sequences (IES). piRNAs generally mark the retained MDS regions (Fang et al. 2012), but here, for mutant strain construction, we instead injected synthetic sRNAs that mark and protect IES regions that are normally deleted: either the IES between MDS 4 and 5 of the Dcl-1 gene (which creates a 24-bp insertion that contains a premature stop codon) or the IES between MDS 6 and 7 of the RdRP gene (which leads to a 52-bp insertion and a resulting frameshift). Exconjugant cells (zygotes) were singled out in 24-well plates and allowed to grow for 7–10 d. PCR across the aforementioned MDSs identified clones that retain the sequences as somatic DNA insertions in the progeny MAC. Positive clones were expanded, encysted, and stored at −80°C.
DNA, RNA isolation, and Illumina library preparation
Whole-cell genomic DNA was isolated from either the JRB310 wild-type strain, the wild-type sexual progeny (exconjugants) or dcl-1 mutants, dcl-123 and dcl-141, and rdrp mutants, rdrp13, rdrp17, and rdrp25 using the Nucleospin Tissue DNA Extraction kit (Macherey-Nagel). One microgram of total genomic DNA was used to prepare Illumina libraries using a standard manufacturer's protocol and run on Illumina HiSeq 2500. Small RNAs were gel-purified from total RNA and loaded on denaturing 7 M Urea, 15% polyacrylamide gel. Of note, 17- to 25-nt small RNAs or 30- to 50-nt RNA (from dcl-141) were gel purified and cloned using the Illumina Truseq small RNA library construction kit. For RNA-seq, total RNA was isolated from wild-type reference strain JRB310 and two mutant strains, dcl-141 and rdrp13. Poly(A) RNA enrichment was performed using the polyA Spin mRNA Isolation Kit (NEB). Enriched RNA was reverse transcribed using Superscript III (ThermoFisher) and strand-specific libraries were constructed using published methods (Zhang et al. 2012). Barcoded libraries were mixed and sequenced.
Sequence analysis
Most of the sequence preprocessing was performed using programs available on the Galaxy tool on the Princeton University webserver (galaxy.princeton.edu) (Goecks et al. 2010). For genomic DNA sequencing, the sequences from exconjugants or dcl-141 were separated based on the barcode, allowing one mismatch, using Barcode Splitter from the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/index.html). Lower quality reads (<Q20) were discarded and the adapter was clipped using Cutadapt (Martin 2011). Adapters were clipped from the 3′ end allowing a 0.1 error rate and a minimum overlap of eight bases. Redundant reads were collapsed using FASTA Collapser from the FASTX-Toolkit to get rid of potential PCR duplicates. For mapping, a master database was prepared including Oxytricha MAC, mitochondrial genome assemblies, common bacterial contaminants and Chlamydomonas reinhardtii. Bowtie2 (v2.2.3) was used for mapping using extra sensitive and end-to-end options (Langmead and Salzberg 2012). Unmapped reads were then aligned to the MIC genome assembly to identify MIC-limited sequences. Coverage was calculated using SAMtools (Li et al. 2009). The wild-type and mutant data sets were normalized by the number of reads mapped per million per kilobase (RPKM). The data sets were further normalized by the number of reads mapping to the mitochondrial genome in each sample.
Small RNA sequence reads were processed in a similar fashion. In addition, reads of length 17–25 nt or 30–50 nt (for sRNA and pre-sRNA, respectively) were filtered for mapping. Pie charts comparing small RNA mapping with genome annotation involved mapping on fully assembled two telomere MAC contigs from Oxytricha strains JRB310 and JRB510 (Swart et al. 2013; Chen et al. 2015). For metagene analysis, two-telomere MAC contigs were aligned at the transcription start sites (TSS). Single-gene chromosomes were normalized to 1 kb in length and used for mapping strand-specific sRNA reads. Single- and two-gene chromosomes were also aligned at stop codons, without length normalization, to compare sRNA mapping 200-bp upstream and downstream from the TSS versus stop codons.
To infer the genomic source of small RNAs, the reads were mapped to MDS–MDS junctions (which can only derive from the MAC) as well as to MDS–IES junctions (which are specific to the MIC). Intersect program from BEDTools Suite (v2.24) was used for the analysis (Quinlan and Hall 2010). Of the 9,009,681 nonredundant small RNA reads, 5,538,289 reads map to the two telomere containing MAC assembly, of which 237,160 reads (4.28%) completely overlap MDS–MDS junctions. Three million 21-nt, 5′-U sequences were generated from the same two-telomere containing MAC assembly using DNemulator (http://www.cbrc.jp/dnemulator) and mapped back to the completely sequenced (two-telomere containing) contigs. Of this randomized set of 21-nt words, 118,933 (3.87%) map to the MDS–MDS junctions. On the other hand, 21,134 sRNA reads (0.31%) map to MDS–IES junctions, whereas 1.25% of a set of similarly produced, randomly generated 21 nt, 5′U sequences from the complete MIC assembly overlap MDS–IES junctions. The published MAC and MIC genome assemblies were used for this analysis (Swart et al. 2013; Chen et al. 2014). A similar analysis was done for the 30- to 50-nt sRNAs, which also primarily map to MAC-specific regions, as shown in Supplemental Figure S3E.
Figure 2C, showing small RNA mapping on individual MAC contigs, was prepared using the Integrated Genomics Viewer (www.broadinstitute.org/igv/home). Length histogram and box plots were prepared in MATLAB. Sequence logos for 21-nt sRNAs were prepared using Weblogos 3.3 (Crooks et al. 2004). Random 21-nt words from the MAC were generated using DNemulator. MIC genome maps for the dcl-1 and rdrp gene loci showing the sequences of piRNAs injected (Supplemental Fig. S2) were prepared in Geneious (http://www.geneious.com/).
Pairwise comparisons of sRNA reads mapping to 12,203 complete MAC chromosomes were performed by creating boxplots in MATLAB, and Wilcoxon rank sum tests were performed between WT and mutant strains to test for statistical significance.
Immunostaining
Cells were treated as described previously (Khurana et al. 2014). Anti-BrdU antibody (1:200) (MBL International) was used for detection of BrdU incorporated into newly replicated DNA. Fixed cells were labeled with the antibody, with or without acidic denaturation. For BrdU signal quantification, we calculated total cell corrected fluorescence (TCCF) using Fiji, as described before (Schindelin et al. 2012; McCloy et al. 2014). Briefly, a target area from BrdU channel was chosen and signal intensity was calculated using Fiji. Similarly, an area adjacent to the positive signal was used to calculate the background signal. TCCF was calculated as the intensity density − (area × mean of background).
Cellular fractionation
Cells were collected and the cell pellet was resuspended in lysis buffer I (10 mM HEPES pH 7.4, 10 mM KCl, 0.05% NP-40) and incubated on ice for 20 min. Cells were then centrifuged at 16,000 rpm for 10 min and the supernatant was collected as the cytoplasmic fraction. The nuclear pellet was washed once in lysis buffer I, followed by resuspension in lysis buffer II (10 mM Tris–HCl pH 7.4, 2 mM MgCl2, 1% Triton-X 100), and then centrifugation at 16,000 rpm for 10 min. The supernatant was collected as nucleoplasm and the pellet (chromatin fraction) was resuspended in a buffer containing 50 mM sodium acetate (pH 5.5), 50 mM NaCl, 0.5% SDS, and vortexed to dissolve completely. For RNA purification, the fractions were subjected to Proteinase K and DNase digestions in consecutive reactions. The RNA was radiolabeled and loaded on denaturing polyacrylamide gels, as described earlier. For protein purification, the fractions were run through 3K Amicon centrifuge columns (Millipore), resuspended in SDS loading buffer and run on Tris–glycine SDS polyacrylamide gels. The gel was blotted onto PVDF membrane (Bio-Rad) and hybridized via Western analysis to an Anti-Histone H3 antibody (Cell Signaling).
RT-PCR and qPCR analysis
Reverse-transcriptase PCR (RT-PCR) was performed using Superscript III per manufacturer's instructions (Invitrogen). Quantitative PCR (qPCR) was performed using Power SyBR green master mix (Life Technologies) and the data were analyzed using either the ddCt method or absolute quantification, as described previously (Swart et al. 2013). For DNA copy-number analysis, an equal number of cells were used for genomic DNA isolation and equal volumes of DNA were used for different samples. Error bars represent standard deviation from an average of three technical replicates from three biological replicates.
dsRNA feeding and sRNA injection
E. coli HT115 cells were transformed with the dsRNA producing vector, L4440, either empty or expressing cDNA portions of Cbx5, Dcl-2, SetD3, and Ku80 genes. Oxytricha cells were fed heat-killed, dsRNA-expressing bacteria, as described previously (Nowacki et al. 2009). DNA and RNA were isolated from cells and analyzed 4 d after dsRNA feeding. Three synthetic duplexed sRNA oligonucleotides with (IDT) against Contig22903.0 were injected in asexually growing cells. Four to five lines were clonally expanded for 4 d post injection, after which DNA was extracted for copy number analysis.
Primer sequences used in this study
DNA copy number and RT-qPCR analysis
SetD3-F: GCAGCATGCACTCCTCTGTA
SetD3-rev: TTCGAGACGCTGTTGAAGTG
Ku80-F: TCTGAGCTTCGTCCTCAGTG
Ku80-rev: ACAGTCTGAACGGGGACTTG
Dcl2-F: CATCGTCCTCAGTGAAGCAA
Dcl2-rev: CTTAGGTGGCTGGCAAAGAG
Cbx5f: TGGAGCAGTAAGTACCATTGGAAG
Cbx5-rev: TGGTTAGGTGCTTCCTTATCTTGA
Contig23093-F: CGTCAACGAAGAGGGAGTTT
Contig23093-rev: CTTCGTCTCCTTCCTCCTCA
Mutant strain confirmation
Dcl1-MDS4-F: CCTTCTCCTGCCTTCTTGTCA
Dcl1-MDS5-rev: GAGCTTGTCAAATGCCCTAAGTG
RdRP-MDS7-F: TTGACCTCTTCTGGCAAACA
RdRP-MDS6-rev: TCCATTTTGGAAGTATGAAGCA
Duplexed sRNAs against target chromosomes (showing sense strand)
Contig23093.0
rUrArCrArArArArArArCrArUrArUrCrUrArArCrArU
rUrGrArCrUrGrArUrCrArGrUrArUrCrCrUrCrUrGrU
rUrGrArArGrGrCrGrUrCrArArCrGrArArGrArGrGrG
Contig13337.0
rUrGrArArGrArUrUrGrArUrArUrArArArUrUrCrArC
rUrArGrArCrCrUrGrCrUrUrCrArArArArGrArUrGrC
rUrArGrGrArGrArUrUrArGrUrUrUrArArGrArArUrU
GFP
rUrGrUrCrUrCrCrGrGrUrGrArArGrGrUrGrArArGrG
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank the current and past members of the Landweber laboratory at Princeton and Columbia for their useful insights and suggestions, particularly Xiao Chen for bioinformatics advice and Jingmei Wang for laboratory support. We are also grateful to Wei Wang and Jessica Buckles Wiggins from the Sequencing Core Facility at Princeton University for Illumina sequencing. The raw and processed sequence files have been submitted to the NCBI sequence archive under the accession number GSE104078. This work was funded by National Institutes of Health grants GM59708, GM111933, and Human Frontier Science Program RGP004/2014.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.061333.117.
Freely available online through the RNA Open Access option.
REFERENCES
- Axtell MJ. 2013. Classification and comparison of small RNAs from plants. Annu Rev Plant Biol 64: 137–159. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366. [DOI] [PubMed] [Google Scholar]
- Castel SE, Martienssen RA. 2013. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet 14: 100–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel SE, Ren J, Bhattacharjee S, Chang AY, Sánchez M, Valbuena A, Antequera F, Martienssen RA. 2014. Dicer promotes transcription termination at sites of replication stress to maintain genome stability. Cell 159: 572–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Bracht JR, Goldman AD, Dolzhenko E, Clay DM, Swart EC, Perlman DH, Doak TG, Stuart A, Amemiya CT, et al. 2014. The architecture of a scrambled genome reveals massive levels of genomic rearrangement during development. Cell 158: 1187–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Jung S, Beh LY, Eddy SR, Landweber LF. 2015. Combinatorial DNA rearrangement facilitates the origin of new genes in ciliates. Genome Biol Evol 7: 2859–2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvillion MT, Lee SR, Hogstad B, Malone CD, Tonkin LA, Sachidanandam R, Hannon GJ, Collins K. 2009. Sequence, biogenesis, and function of diverse small RNA classes bound to the Piwi family proteins of Tetrahymena thermophila. Genes Dev 23: 2016–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14: 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Malone C, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel J, Sachidanandam R, et al. 2008. An endogenous small interfering RNA pathway in Drosophila. Nature 453: 798–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC. 2000. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101: 543–553. [DOI] [PubMed] [Google Scholar]
- Fang W, Wang X, Bracht JR, Nowacki M, Landweber LF. 2012. Piwi-interacting RNAs protect DNA against loss during Oxytricha genome rearrangement. Cell 151: 1243–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley BM, Collins K. 2017. Transgenerational function of Tetrahymena Piwi protein Twi8p at distinctive non-coding RNA loci. RNA 23: 530–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. [DOI] [PubMed] [Google Scholar]
- Fujiu K, Numata O. 2000. Reorganization of microtubules in the amitotically dividing macronucleus of Tetrahymena. Cell Motil Cytoskeleton 46: 17–27. [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore P. 2009. Small silencing RNAs: an expanding universe. Nat Rev Genet 10: 94–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler ELW, Zapp ML, Weng Z, et al. 2008. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 320: 1077–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J; Galaxy Team. 2010. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11: R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404: 293–296. [DOI] [PubMed] [Google Scholar]
- Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada T, Siomi M, Siomi H. 2008. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature 453: 793–797. [DOI] [PubMed] [Google Scholar]
- Khurana JS, Wang X, Chen X, Perlman DH, Landweber LF. 2014. Transcription-independent functions of an RNA polymerase II subunit, Rpb2, during genome rearrangement in the ciliate, Oxytricha trifallax. Genetics 197: 839–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC, Chang SS, Choudhary S, Aalto AP, Maiti M, Bamford DH, Liu Y. 2009. qiRNA is a new type of small interfering RNA induced by DNA damage. Nature 459: 274–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepère G, Nowacki M, Serrano V, Gout JF, Guglielmi G, Duharcourt S, Meyer E. 2009. Silencing-associated and meiosis-specific small RNA pathways in Paramecium tetraurelia. Nucleic Acids Res 37: 903–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad KA, Bracht JR, Williams AE, Landweber L. 2017. Thousands of RNA-cached copies of whole chromosomes are present in the ciliate Oxytricha during development. RNA 23: 1200–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17: 10–12. [Google Scholar]
- McCloy RA, Rogers S, Caldon CE, Lorca T, Castro A, Burgess A. 2014. Partial inhibition of Cdk1 in G2 phase overrides the SAC and decouples mitotic events. Cell Cycle 13: 1400–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, et al. 2007. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA. 2002. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in Tetrahymena. Cell 110: 689–699. [DOI] [PubMed] [Google Scholar]
- Nowacki M, Vijayan V, Zhou Y, Schotanus K, Doak TG, Landweber LF. 2008. RNA-mediated epigenetic programming of a genome-rearrangement pathway. Nature 451: 153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowacki M, Higgins BP, Maquilan GM, Swart EC, Doak TG, Landweber LF. 2009. A functional role for transposases in a large eukaryotic genome. Science 324: 935–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowacki M, Haye JE, Fang W, Vijayan V, Landweber LF. 2010. RNA-mediated epigenetic regulation of DNA copy number. Proc Natl Acad Sci 107: 22140–22144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Chung W-J, Ruby JG, Guo H, Bartel D, Lai E. 2008. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature 453: 803–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Karpen GH. 2007. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat Cell Biol 9: 25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips DD, Garboczi DN, Singh K, Hu Z, Leppla SH, Leysath CE. 2013. The sub-nanomolar binding of DNA–RNA hybrids by the single-chain Fv fragment of antibody S9.6. J Mol Recognit 26: 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott DM. 1994. The DNA of ciliated protozoa. Microbiol Rev 58: 233–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A. 2001. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107: 465–476. [DOI] [PubMed] [Google Scholar]
- Swart EC, Nowacki M, Shum J, Stiles H, Higgins BP, Doak TG, Schotanus K, Magrini VJ, Minx P, Mardis ER, et al. 2012. The Oxytricha trifallax mitochondrial genome. Genome Biol Evol 4: 136–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swart EC, Bracht JR, Magrini V, Minx P, Chen X, Zhou Y, Khurana JS, Goldman AD, Nowacki M, Schotanus K, et al. 2013. The Oxytricha trifallax macronuclear genome: a complex eukaryotic genome with 16,000 tiny chromosomes. PLoS Biol 11: e1001473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al. 2008. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453: 534–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terradas M, Martín M, Tusell L, Genescà A. 2010. Genetic activities in micronuclei: is the DNA entrapped in micronuclei lost for the cell? Mutat Res 705: 60–67. [DOI] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Theurkauf WE, Weng Z, Zamore PD. 2012. Strand-specific libraries for high throughput RNA sequencing (RNA-Seq) prepared without poly(A) selection. Silence 3: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.