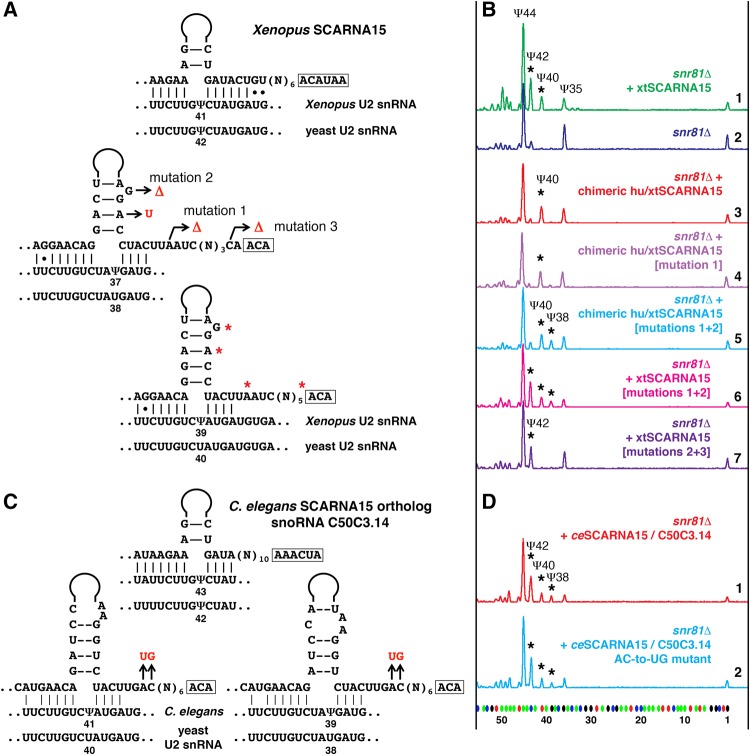
FIGURE 3.
Pseudouridylation of yeast U2 snRNA induced by Xenopus SCARNA15 and by the C. elegans ortholog, snoRNA C50C3.14. (A,C) Predicted base-pairing between SCARNA15 and Xenopus U2 snRNA (A) or C. elegans U2 snRNA (C). The equivalent yeast U2 snRNA sequences are shown below. Mutated nucleotides are highlighted in red. (B,D) Fluorescent primer extension reactions. (B) Traces 1 (green) and 2 (blue): Wild-type Xenopus SCARNA15 modifies yeast U2 snRNA at positions 42 and 40 when expressed in the snr81Δ mutant yeast strain. Trace 3 (red): Chimeric SCARNA15 (human 5′ terminal hairpin and Xenopus 3′ terminal hairpin) modifies only position 40 of yeast U2 snRNA. Trace 4 (violet): Mutation 1 alone did not change pseudouridylation of position 38. Traces 5 (light blue) and 6 (magenta): A combination of mutations 1 and 2 is required to make the Xenopus 3′ terminal pseudouridylation pocket functional at position 38 in U2 snRNA. Trace 7 (dark violet): Mutation 3 makes the 3′-terminal domain of Xenopus SCARNA15 nonfunctional on yeast U2 snRNA; only the 5′ terminal pocket functions to position Ψ42. Stars indicate induced modifications. (D) Traces 1 (red) and 2 (blue): Both wild-type (red) and mutant C. elegans SCARNA15 (blue) are fully functional at all three predicted positions in yeast U2 snRNA: Ψ42, Ψ40, and Ψ38 (stars).