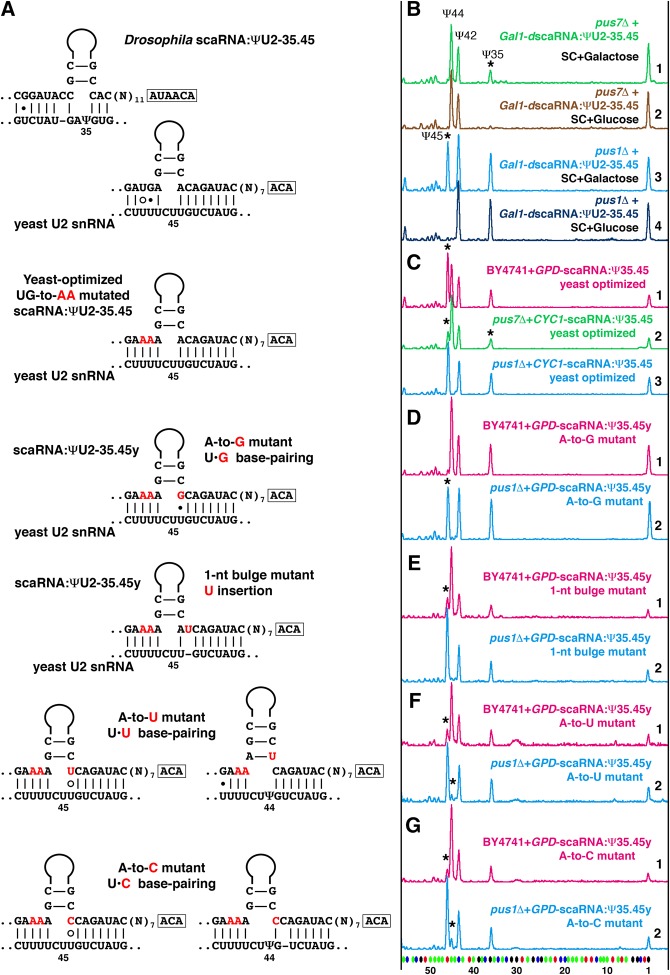
FIGURE 5.
Pseudouridylation of U2 snRNA at position 44 interferes with pseudouridylation at position 45 in the yeast cell system. (A) Base-pairing of wild-type and mutated variants of Drosophila scaRNA:ΨU2-35.45 with yeast U2 snRNA. Mutated nucleotides are shown in red. (B–G) Fluorescent primer extension reactions. (B) Traces 1 (green) and 2 (light brown): Drosophila scaRNA:ΨU2-35.45 cannot induce pseudouridylation at position 45 when expressed stepwise from an inducible Gal promoter in the pus7Δ yeast strain. Rescue of pseudouridylation at position 35 (star) serves as an internal positive control. Traces 3 (blue) and 4 (dark blue): In the same experimental setup, Drosophila scaRNA:ΨU2-35.45 is active at position 45 (star) in the pus1Δ strain, which has no Ψ44 in U2 snRNA. (C) Trace 1 (magenta): scaRNA:ΨU2-35.45 optimized for positioning Ψ45 in yeast U2 snRNA efficiently modifies position 45 in wild-type yeast strain only when overexpressed. Trace 2 (green): The same yeast-optimized RNA modifies position 45 inefficiently in Pus1p-positive strains, such as pus7Δ. Trace 3 (blue): This RNA is fully functional at position 45 in the pus1Δ strain. (D–G) Traces 1 (magenta): Point mutations in the 3′ side of the yeast-optimized U2-Ψ45 pseudouridylation pocket affect the corresponding modification activity, especially in the presence of U2-Ψ44 in a “wild-type” yeast strain. Traces 2 (blue): The U2-Ψ45 modification activity is evident in the absence of U2-Ψ44 in the pus1Δ yeast strain. Stars indicate guide RNA-induced pseudouridines.