FIGURE 8.
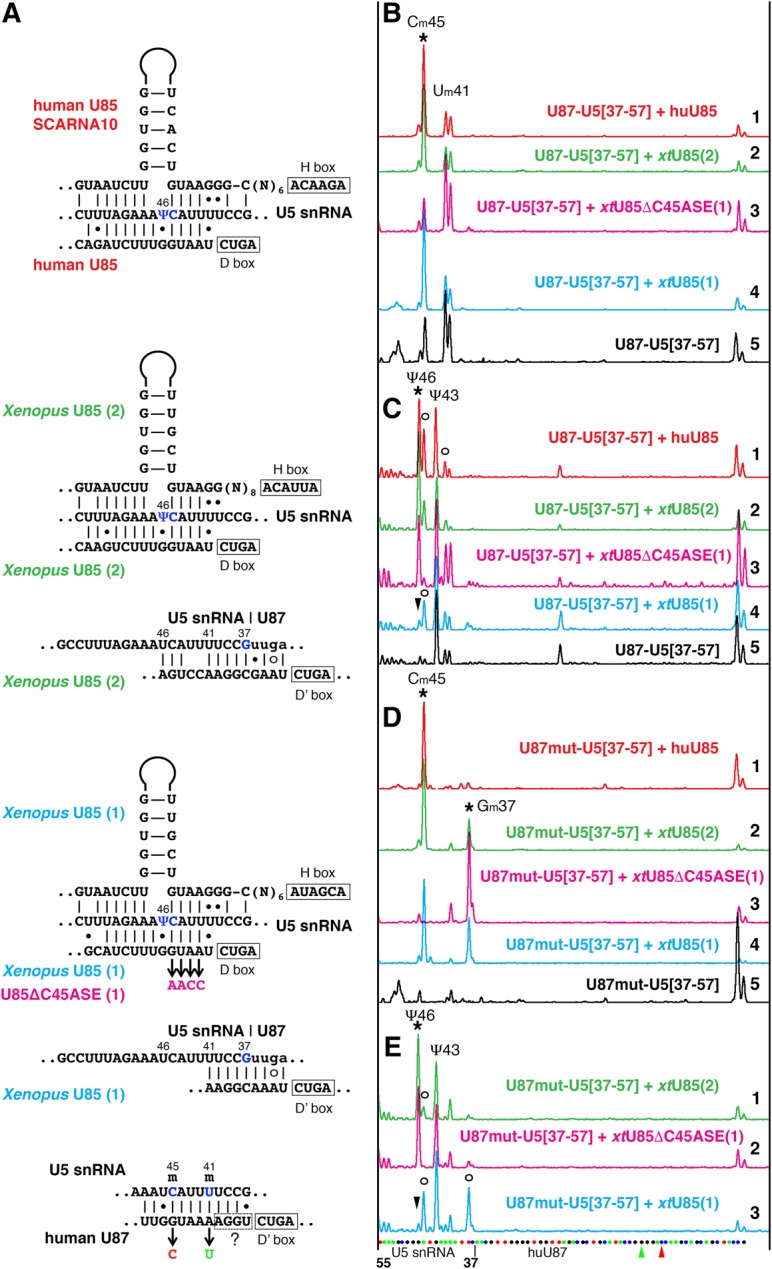
U85 scaRNA modification activity on U5 snRNA. (A) Predicted interactions of human U85, Xenopus U85, and human U87 scaRNAs with U5 snRNA. Targeted positions are shown in blue. Mutations introduced in antisense elements of Xenopus U85 and human U87 RNAs are color-coded to match colors used in B–E. (B–E) Fluorescent primer extension reactions. (B,D) 2′-O-methylation and (C,E) pseudouridylation of the 5′ loop of vertebrate U5 snRNA (nucleotides 37–57) inserted into U87 RNA and expressed in wild-type yeast cells. Stars indicate peaks that correspond to modifications induced by human U85 (traces 1, red in B–D) and two Xenopus wild-type U85 (traces 2, green and 4, blue in B–D; traces 1, 3 in E), and Xenopus U85 with mutated Cm45 antisense element, U85ΔC45ASE (traces 3, magenta in B–D; trace 2 in E). Arrowheads point to missing Ψ46. Trace 5, black in B, C, and D: Control samples of U87–U5 substrate RNAs expressed alone in yeast cells. In the tested artificial U5 snRNA substrate, U87–U5[37–57], position 43 is pseudouridylated by an unknown endogenous yeast snoRNA (C,E) and positions 41 and 45 are 2′-O-methylated by U87 RNA itself (B). (D) When the antisense element for positioning Um41 and Cm45 was removed from U87, U87mut-U5[37–57], an additional position became 2′-O-methylated by Xenopus U85; human U85 could not induce this modification. Open circles in C and E indicate stops at 2′-O-methylated positions. In the interest of space, only three Xenopus U85 traces are shown in E; the pattern induced by human U85 (not shown) is very similar to that in trace 1 (green) of Xenopus U85(2); the control U87mut-U5[37–57] alone (not shown) looks as expected, with one peak at Ψ43 and no extra peak at Um41. The sequence of U87–U5 substrate RNA and mutated positions are shown at the bottom of E.
