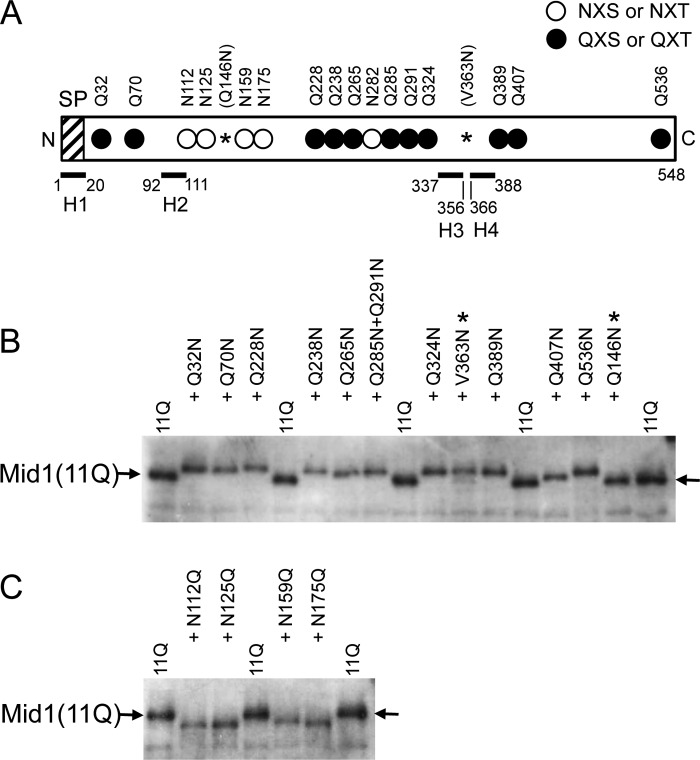
Figure 7.
All putative N-glycosylation sites examined are N-glycosylated. A, schematic diagram of the Mid1(11Q) protein, the 11 putative N-glycosylation sites (Asn) of which were replaced with Gln. Open circle, intact N-glycosylation site; closed circle, mutated N-glycosylation site with the Asn → Gln substitution; *, a newly substituted Asn residue, which produced the Q146N or V363N substitution. Note that the latter generated a new N-glycosylation site, whereas the former did not. SP, signal peptide. H1–H4, the top four hydrophobic regions in Mid1 (7). Note that among the 16 putative N-glycosylation sites, only Asp282 (one of the open circles) was not tested for N-glycosylation because this residue was expected to receive the same modification as the 6 Asn residues near it. In addition, the reason for constructing Q285N/Q291N was that the two positions were so close to each other that it was unnecessary to construct the corresponding single substitution mutants because the purpose of this experiment was to assess the membrane topology of Mid1. B, a putative N-glycosylation site added to the Mid1(11Q) protein decreases its electrophoretic mobility, indicating that it is N-glycosylated. *, newly substituted Asn residue; arrow, the position of Mid1(11Q) on the Western blot. C, the Asn → Gln substitution at a putative N-glycosylation site in the Mid1(11Q) protein increases its electrophoretic mobility, indicating that the substituted site is originally an N-glycosylation site. In B and C, crude extracts prepared from cells expressing each substitution mutant protein were subjected to SDS-PAGE and a Western blot analysis using affinity-purified rabbit polyclonal antibodies against 20 C-terminal amino acid residues of Mid1 (10).