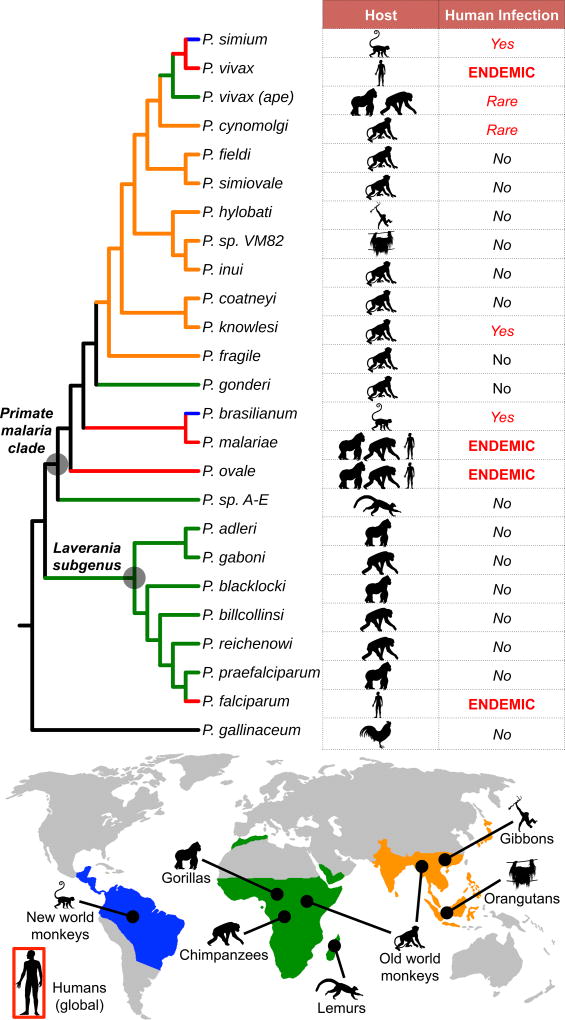
Figure 1. Evolutionary relationships of primate malaria parasites.
Neighbor-joining tree was derived from a 3486-bp gapped mtDNA alignment encompassing the cytochrome oxidase subunit 3, cytochrome oxidase subunit 1-like, and cytochrome b genes of 24 previously published non-human primate malaria parasite sequences and outgroup P. gallinaceum (GenBank accession numbers: AB354571-AB354575, AB434918, AB434919, AB444136, AB489194, AJ251941, AY791692, AY800109, AY800110, EU880499, FJ895307, GQ355477, GQ355484, HM235302, HM235369, HM235379, HQ712054, JF923761, JQ308531, and NC_008288). Lineages are colored corresponding to geographic origin of host species (blue=South America; green=Africa; orange=Asia). Human-adapted lineages, which are globally distributed, are colored red. Lineages are labeled according to host species and degree of specificity for the human host in natural settings.