Figure 1.
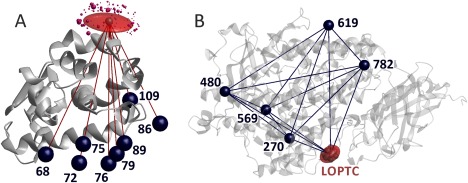
Localization of paramagnetic centers by distance distribution measurements. (A) MTSL at residue 131 in T4 Lysozyme (PDB 2LZM) is localized by multilateration using eight distances to other spin‐labeled residues and the standard deviations of their distributions. Underlying data courtesy H. Mchaourab34 and C. Altenbach (unpublished). Dark blue spheres visualize the mean position of the reference spin sites, the red semi‐transparent surface includes 50% of the probability for finding the spin at site 131, and purple spheres the spin density centers of MTSL rotamers predicted by MMM with sphere volume proportional to their predicted population. (B) LOPTC in soybean seed lipoxygenase‐1 (PDB 1YGE) is localized by distance geometry, taking into account distances and their standard deviations to five reference sites labeled with MTSL as well as the ten distances between the reference sites.31 The red semi‐transparent surface includes 50% of the probability for finding the spin of the labeled lipid
