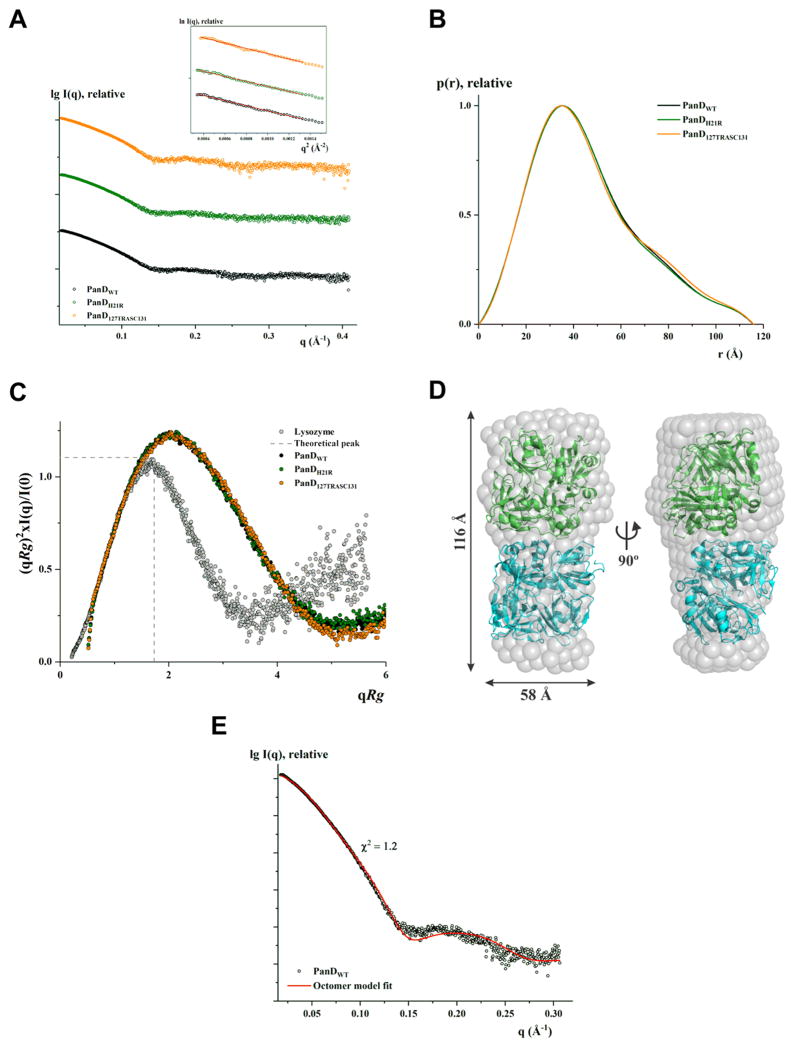
Figure 6.
Solution X-ray scattering studies of PanD proteins. (A) SAXS pattern (○) of WT PanD (PanDwt) at 7.4 mg/mL (black), PanDH21R at 5.0 mg/mL (green), and PanD127TRASC131 at 6.5 mg/mL (orange) concentrations. (Inset) Guinier plots show linearity at the concentrations used, indicating no aggregation. The scattering profiles are offset for clarity by applying arbitrary scale factors. (B) Overlapping of pair-distance distribution function P(r) of PanDwt (—; black), PanDH21R (—; green), and PanD127TRASC131 (—; orange) show no difference. (C) Normalized Kratky plot of PanDwt (●; black) compared to the mutants PanDH21R (●; green) and PanD127TRASC131 (●; orange) and the compact globular lysozyme (●; gray) with a peak (---; gray), representing the theoretical peak and assuming an ideal Guinier region of a globular particle. The scattering pattern of wild-type and mutants of PanD are similar and exhibits a broad bell-shaped profile shifted toward the right with respect to standard globular protein. (D) The averaged and filtered ab initio DAMMIN envelope (NSD = 0.47 ± 0.01) of WT PanD (gray spheres) superimposed onto the cartoon representation of the octameric model of PanD generated using the tetrameric crystallographic structure (PDB ID: 2C45). The tetrameric subunits are colored green and cyan. Front (left) and side (right) views are displayed. (E) Fitting of the experimental scattering pattern of WT PanD (○; black) and calculated scattering profile of the octameric model (—; red) derived from crystal structure using SASREF.