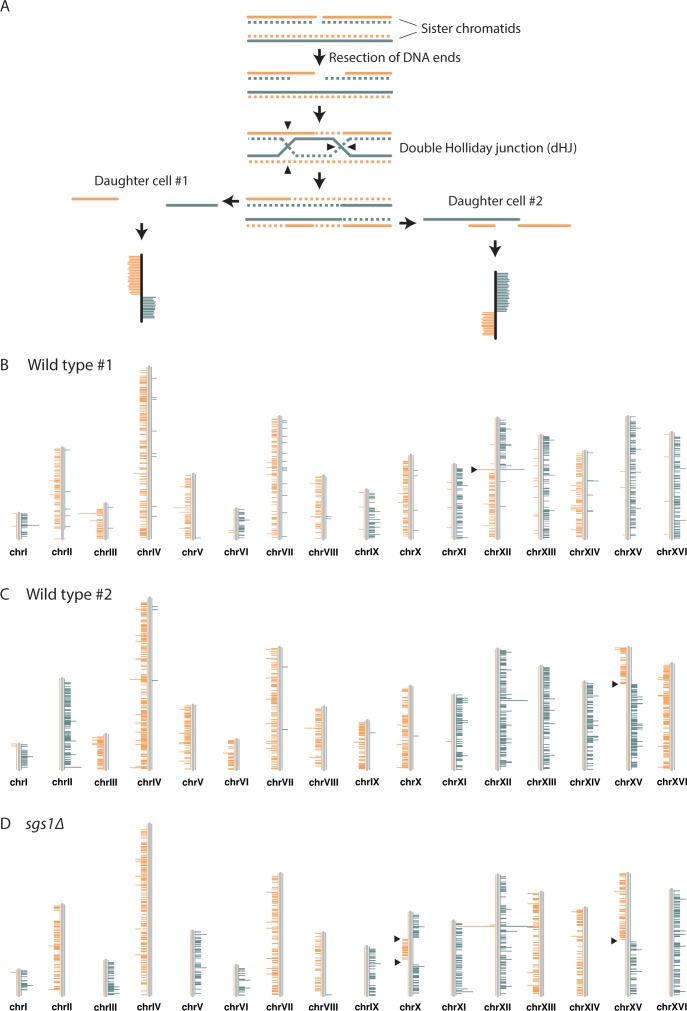
Figure 1. Detection of SCE events using Strand-seq.
(A) An SCE can occur as a result of DSB repair. Two sister chromatids, one of which has a DSB, are shown. The parental template DNA strands are depicted with solid lines, while the newly synthesized strands containing BrdU are depicted with dashed lines. The Watson and Crick strands are shown in orange and blue, respectively. DSB repair by SCR can lead to the formation of a double Holliday junction (dHJ). Resolution of the dHJ by structure-specific endonucleases will result in either a noncrossover (not shown) or a crossover. The resulting sister chromatids are then segregated to two different daughter cells. In the current Strand-seq protocol, only one daughter cell is isolated and analyzed. The BrdU-containing strands are nicked during library preparation, resulting in the sequencing of only parental strands. Sequence reads are mapped to either side of a chromosome ideogram. An SCE results in a switch from Watson to Crick reads along the chromosome. Note: the small gap between the parental strands in daughter cell #1 and the small overlap of the parental strands in daughter cell #2 are too small to be detected with Strand-seq. (B) An example of a wild-type Strand-seq library. Ideograms of the 16 yeast chromosomes are shown. Orange and blue lines correspond to reads aligning to the Watson and Crick strands, respectively. This cell inherited either the parental Watson strand or the parental Crick strand for each chromosome, except chromosome XII. A switch from Watson to Crick reads can be seen for chromosome XII (black arrowhead), indicating that an SCE event has occurred. (C) A second example of a wild-type Strand-seq library. An SCE event was detected on chromosome XV. (D) An example of an sgs1∆ Strand-seq library. Three SCE events were detected in this library: two on chromosome X and one on chromosome XV.