Abstract
Localization of specific proteins within cells at the nanometer level of resolution is central to understanding how these proteins function in cell processes such as motility and intracellular trafficking. Such localization can be achieved by combining transmission electron microscopy (TEM) with immunogold labeling. Here we describe a pre-embedding, indirect gold immunolabeling approach to localize two different proteins of interest with secondary antibodies labeled with gold particles of different sizes in cells grown on cover slips. In this protocol, the cells are immunolabeled prior to being embedded in an epoxy resin for ultrathin sectioning. The protocol also includes strategies for optimizing the balance between ultrastructure and antigen preservation, steps to minimize nonspecific antibody binding, and steps to optimize antibody penetration.
Keywords: Immunogold, Antibodies, Monolayer, Pre-embedding
I Introduction
Since Faulk and Taylor [1] took advantage of the high electron density of gold to map the distribution of Salmonella antigens by transmission electron microscopy (TEM) and Frens [2] developed methods to control the size of colloidal gold particles, several thousand studies have used variations of these basic protocols to visualize, at the ultrastructural level, the localization of specific proteins in cells and tissues. Combined with the results of molecular, biochemical, and fluorescence light microscopy approaches, the results of these ultrastructural studies have contributed to our understanding of the function of numerous proteins. In fact, immunogold electron microscopy is the only morphological method with sufficient spatial resolution to provide definitive localization of proteins within organelles such as lysosomes, mitochondria, or synapses (e.g., [3–5]) or the association of protein linkers with organelles or cytoskeletal polymers (e.g., [5–9]).
While immunogold TEM on tissues enables investigators to determine protein localization in situ, transfection of cultured cells enables them to localize protein domains or tagged proteins and to use correlative microscopy to determine the distribution of specific proteins in the same cell at the light and electron microscope levels [10, 11]. Another advantage of using cultured cell monolayers is that they are often thinner than tissue sections, thereby facilitating antibody penetration. The protocol presented in this chapter considers only cultured cell monolayers since 3D cultures, depending on their thickness, may be more similar to tissue sections and should be processed as such for immunogold labeling for TEM. Cultures of non-adherent, floating cells can also be processed for immunogold TEM using protocols developed for tissue sections after collecting these non-adherent cells as a pellet by centrifugation [12]
Immunolabeling can be direct or indirect. In direct immunolabeling, the primary antibody is conjugated to gold particles. In indirect immunolabeling, the primary antibody is unconjugated and labeling is achieved with a gold-conjugated secondary antibody specific for the primary antibody. The advantage of the indirect procedure is that multiple secondary antibodies can bind to the primary antibody increasing the signal. The disadvantage is that the gold particles are further away (15–30 nm) from the site the primary antibody binds to on the antigen.
Immunolabeling can also be performed pre- or post-embedding. In post-embedding protocols, the cells are fixed with a strong cross-linking chemical, usually glutaraldehyde, then embedded in epoxy or low-viscosity resin and sectioned with an ultramicrotome. The sections obtained are then immunolabeled. In pre-embedding protocols, the cells are fixed with a weak fixative, usually formaldehyde, followed by immunolabeling. The cells are then embedded in epoxy resin and sectioned with an ultramicrotome. Post-embedding protocols using cells embedded in epoxy resin offer excellent ultrastructural preservation but often result in poor labeling because protein epitopes are altered by the glutaraldehyde fixation and the high temperature associated with the curing of the epoxy resin during embedding. This results in reduced primary antibody binding and thus a lower signal. Pre-embedding protocols offer better epitope preservation, better antibody binding, and thus a stronger signal. The trade-off of the pre-embedding protocol is reduced ultrastructural preservation because the fixatives used are weaker than those used in post-embedding protocols.
The pre-embedding protocol described in this chapter utilizes a post-labelling fixation step to minimize ultrastructural loss. In this step, immunolabeled cells are subsequently fixed with glutaraldehyde and osmium in order to stabilize the antibody complexes and to prevent ultrastructural degradation that occurs during the dehydration and heating steps associated with embedding in epoxy resins. This methodology for indirect double immunolabeling allows the distribution and localization of two proteins to be determined simultaneously. Combined with biochemical and genetic analysis, double immunolabeling provides another method to demonstrate the interaction of two proteins and to locate the cellular regions or organelles where this interaction occurs [6, 9].
2 Materials
2.1 Plating Cells on Cover Slips
Human microvascular endothelial cells (Cascade Biologics, Portland, OR) (see Note 1).
Tissue culture medium: Medium 131 supplemented with microvascular growth supplement (MVGS) (Thermo Fisher Scientific, Grand Island, NY).
Sterile trypsin-EDTA: 0.25% Trypsin, 2.21 mM EDTA (Thermo Fisher Scientific, Grand Island, NY).
Fetal bovine serum (FBS) (Thermo Fisher Scientific, Grand Island, NY).
Trypsin inhibitor medium: Mix 90 mL Medium 131 and 10 mL FBS in a sterile capped bottle or tube.
13 mm Diameter sterile Thermanox plastic cover slips (Thermo Fisher Scientific, Grand Island, NY) (see Note 2).
60 mm Sterile plastic culture dishes.
6-Well sterile plastic tissue culture plates.
Sterile 10 mL plastic or glass pipettes.
15 mL Sterile disposable plastic conical centrifuge tubes.
Pointed-tip tweezers, style 3 (Electron Microscopy Sciences, Hatfield, PA).
Flat-tip tweezers (Electron Microscopy Sciences, Hatfield, PA).
Hemocytometer.
CO2 tissue culture incubator.
Laminar flow biosafety cabinet.
Desktop centrifuge.
Inverted phase-contrast microscope.
2.2 Immunogold Labeling and Post-labeling Fixation of Cells on Cover Slips
10× PBS buffer: Dissolve 80 g of NaCl, 2 g of KCl, 14.4 g of Na2HPO4, and 2.4 g of KH2PO4 in 800 mL of distilled H2O. Adjust the pH to 7.4 with HCl. Make up the volume to 1 L with distilled H2O. Autoclave to sterilize (see Note 3).
1× PBS: Add 100 mL of 10× PBS to 900 mL of distilled water and mix well.
4% Paraformaldehyde fixation solution (see Note 4): Mix 1 g of paraformaldehyde with 20 mL of water, add two drops of 1 N NaOH, heat to 60 °C until powder dissolves (do not allow to boil!), cool, add 2.5 mL of 10× PBS, adjust pH to 7.4 with 1 N NaOH, and add water to a final volume of 25 mL. Make fresh for each procedure.
Free aldehyde group blocking solution: Add 375 mg glycine to 100 mL 1× PBS, mix well, and adjust pH to 7.4 with 0.2 M HCl.
Permeabilization solution: Add 1 mL of 10% Triton-X-100 (EMD Millipore, Billerica, MA) to 99 mL lx PBS and mix well.
Blocking solution: Add 10 mL of normal horse serum to 90 mL of 1× PBS and mix well.
Bovine serum albumin (BSA) (see Note 5).
A primary antibody against each of the two proteins being studied (see Notes 6 and 7). The two primary antibodies must be raised in different species.
Two secondary antibodies conjugated with different size gold particles (Electron Microscopy Sciences, Hatfield, PA) (see Note 8), which can be raised in the same species but that species must be different from that of the two primaries.
Antibody diluent: 1 % Bovine serum albumin (BSA) in 1× PBS.
2.5% Glutaraldehyde in sodium cacodylate buffer (Tousimis, Rockville, MD) (see Note 4).
EM wash buffer (0.1 M sodium cacodylate-HCL pH 7.2): Mix 21.4 g of sodium cacodylate trihydrate (AsO2Na(CH3)2·3H2O) with 900 mL of water, adjust the pH to 7.2 with 0.2 M HCl, and make up to a final volume of 1 L with water.
0.2 M Sodium cacodylate buffer: Mix 21.4 g of sodium cacodylate trihydrate (AsO2Na(CH3)2·3H2O) to 450 mL of water, adjust the pH to 7.2 with 0.2 M HCl, and make up to a final volume of 0.5 L with water.
Osmium post-fix solution (see Notes 4 and 9): Immediately prior to use, mix equal volumes of 4 % aqueous osmium tetroxide (OsO4) solution (Electron Microscopy Sciences, Hatfield, PA) and 0.2 M sodium cacodylate buffer (see Note 10).
Uranyl acetate staining solution: Mix 6.25 g of uranyl acetate powder (Ted Pella, Inc., Redding, CA) (see Note 11) with 100 mL of distilled water in an amber bottle and tightly seal. Sonicate for up to 1 h. Centrifuge at 15,000×g for 5 min prior to use.
High-precision tweezers, style 3 (Electron Mcroscopy Sciences, Hatfield, PA).
100 mL Beaker.
6-Well sterile plastic tissue culture plates.
24-Well sterile plastic tissue culture plates.
Sonicator (Sonicor Instrument Corp., Copiague, NY).
Tabletop centrifuge.
Chemical fume hood.
2.3 Embedding and Preparation of Ultrathin Sections
Ethanol 100% (200 proof) and a graded series of ethanol dilutions (30, 50, 70, 90, and 95%).
Propylene oxide (Electron Microscopy Sciences, Hatfield, PA).
Embedding resin: EMbed 812 Resin kit (Electron Microscopy Sciences, Hatfield, PA) prepared according to the manufacturer’s instructions (see Notes 12 and 13).
Uranyl acetate staining solution: Mix 6.25 g of uranyl acetate powder (Ted Pella, Inc., Redding, CA) (see Note 11) with 100 mL of distilled water in an amber bottle and tightly seal. Sonicate for up to 1 h. Centrifuge at 15,000×g for 5 min prior to use.
Reynold’s lead citrate staining solution: Add 1.33 g of lead nitrate (Electron Microscopy Sciences, Hatfield, PA) and 1.76 g of sodium citrate dihydrate (Electron Microscopy Sciences, Hatfield, PA) to 30 mL of distilled water. Mix for 1 h (it is normal for the solution to become cloudy when sodium citrate is added). Then add 8 mL of 1 N NaOH (solution becomes clear when NaOH is added). Add 12 mL of distilled water to a final volume of 50 mL. Stir for 10 min. The solution may be filtered through a Millipore filter to remove any undissolved material. Do not use solution if it is cloudy.
High-precision tweezers, style 3 (Electron Mcroscopy Sciences, Hatfield, PA).
Jeweler saw (Electron Microscopy Sciences, Hatfield, PA).
Sonicator (Sonicor Instrument Corp., Copiague, NY).
Tabletop centrifuge.
Millipore filters 0.2 mm pore size.
Whatman #1 filter paper.
Parafilm.
Aluminum foil.
Plastic Petri dishes (100 mm in diameter).
Glass Petri dishes (100 mm in diameter).
BEEM capsules, size 00 (Electron Microscopy Sciences, Hatfield, PA).
BEEM capsule press (Electton Microscopy Sciences, Hatfield, PA).
BEEM capsule holder (Electron Microscopy Sciences, Hatfield, PA).
Ultramicrotome (Leica Microsystems, Buffalo Grove, IL) with diamond knife (Electron Microscopy Sciences, Hatfield, PA).
Single-edge razor blades (Electron Microscopy Science, Hatfield, PA).
Binocular dissecting microscope.
Vacuum oven able to reach 60 °C or higher.
Copper grids, 200 mesh, for transmission electron microscopy (Electron Microscopy Sciences, Hatfield, PA) (see Note 14)
Flat plastic embedding molds (Electron Microscopy Sciences Hatfield, PA).
Storage box for copper grids (Electron Microscopy Sciences Hatfield, PA).
Transmission electron microscope (TEM).
3 Methods
3.1 Plating Cells on Cover Slips
Plate 5 × 105 cells in a 60 mm culture dish containing 5 mL of tissue culture medium (see Note 15).
Place the culture dish in a tissue culture incubator set up at 37 °C and 5 % CO2.
Incubate for 3–4 days or until the culture reaches ~80% confluency as determined by daily observation with an inverted phase-contrast microscope.
Remove the medium from the dish with a vacuum line.
Add 2 mL trypsin-EDTA and incubate at 37 °C for 3–5 min or until the cells have detached from the bottom of the dish as determined with an inverted phase-contrast microscope.
As soon as 80% or more of the cells have detached, neutralize the trypsin activity by adding 2 mL of trypsin inhibitor medium to the dish.
Using a sterile pipette, transfer the 4 mL suspension of detached cells to a 15 mL disposable sterile conical plastic centrifuge tube.
Centrifuge at 1000×g for 5 min at room temperature.
Remove the supernatant with a vacuum line taking care not to aspirate the cell pellet.
With a sterile pipette, gently resuspend the cells in 5 mL of tissue culture medium. Avoid excessive foaming or bubbling during the process.
Determine the cell concentration with a hemocytometer (see Note 16) and dilute the cells with tissue culture medium to a final concentration of 2 × 104 cells/mL.
Using the pointed-tip and the flat-tip tweezers (see Note 17), create a bend on one side of the Thermanox plastic cover slip
Use the sterile pointed-tip tweezers to place one cover slip into each well of a 24-well tissue culture plate. Bended side of the cover slips should be facing up such that the cover slip lays flat in the well.
Add 1 mL of cell suspension at 2 × 104 cells/mL onto the center of each cover slip (see Note 18).
Place the culture dish in a tissue culture incubator set up at 37°Cand 5%CO2.
Incubate for 1–2 days or until the culture reaches 80–90% confluency as determined by daily observation with a phase-contrast microscope (see Note 19).
3.2 Immunogold Labeling and Post-labeling Fixation of Cells on Cover Slips
Using a pointed-tip tweezers and keeping track of the side of the cover slip onto which the cells are attached, lift the cover slip using its bended side (“handle”) and wash off excess medium by dipping the cover slip five times into a 100 mL beaker containing 1× PBS (see Note 20).
Transfer the cover slip to a culture dish filled with paraformaldehyde fixation solution and submerge the cover slip, cells facing up, in the solution (see Note 21). Incubate for 30 min.
Transfer the cover slip to a culture dish filled with 1× PBS and submerge the cover slip, cells facing up, in the solution. Wash for 5 min. Repeat twice.
Transfer the cover slip to a culture dish filled with free aldehyde group blocking solution and submerge the cover slip, cells facing up, in the solution (see Note 22). Block for 30 min.
Transfer the cover slip to a culture dish filled with 1× PBS and submerge the cover slip, cells facing up, in the solution. Wash for 5 min.
Transfer the cover slip to a culture dish filled with permeabilization solution and submerge the cover slip, cells facing up, in the solution (see Note 23). Permeabilize for 5 min.
Transfer the cover slip to a culture dish filled with 1× PBS and submerge the cover slip, cells facing up, in the solution. Wash for 5 min.
Transfer the cover slip to a culture dish filled with blocking solution and submerge the cover slip, cells facing up, in the solution. Block for 5 min.
Transfer the cover slip, cells facing up, to a well of a 24-well plate (see Note 24) filled with 300 μL (total volume) of the two primary antibodies, each diluted at the appropriate dilution (see Note 25) in antibody diluent. Incubate for 1–2 h.
Transfer the cover slip to a culture dish filled with 1× PBS and submerge the cover slip, cells facing up, in the solution. Wash for 10 min. Repeat twice.
Transfer the cover slip, cells facing up, to a well of a 24-well plate (see Note 24) filled with 300 μL (total volume) of the two secondary antibodies conjugated to different size gold particles and each diluted at 1:20 in antibody diluent (see Note 26). Incubate for 1–2 h.
Transfer the cover slip to a culture dish filled with 1× PBS and submerge the cover slip, cells facing up, in the solution (see Note 26). Wash for 10 min. Repeat twice.
Transfer the cover slip to a culture dish filled with EM wash buffer and submerge the cover slip, cells facing up, in the solution (see Note 27). Wash for 5 min. Repeat twice.
Transfer the cover slip to a culture dish filled with 2.5 % glutaraldehyde in sodium cacodylate buffer and submerge the cover slip, cells facing up, in the solution. Incubate for 2 h at room temperature or overnight at 4 °C.
Transfer the cover slip to a culture dish filled with EM wash buffer and submerge the cover slip, cells facing up, in the solution. Wash for 10 min. Repeat three times.
Transfer the cover slip to a culture dish filled with osmium post-fix solution and submerge the cover slip, cells facing up, in the solution. Incubate for 2 h in the dark in a fume hood (see Notes 4, 9 and 10).
Transfer the cover slip to a culture dish filled with EM wash buffer and submerge the cover slip, cells facing up, in the solution. Wash for 15 min. Repeat twice.
Transfer the cover slip to a culture dish filled with distilled water and submerge the cover slip, cells facing up, in the solution. Wash for 15 min. Repeat twice.
Transfer the cover slip to a culture dish filled with uranyl acetate staining solution (see Note 11) and submerge the cover slip, cells facing up, in the solution. Stain in the dark for 30 min.
Transfer the cover slip to a culture dish filled with distilled water and submerge the cover slip, cells facing up, in the distilled water. Wash for 5 min. Repeat twice.
3.3 Embedding and Preparation of Ultrathin Sections
Dehydrate the cells by sequentially incubating the cover slip, cell side facing up, in culture dishes filled with a graded series of ethanol: 30% (3×), 50% (3×), 70% (3×), 90% (twice), 95% (twice), and 100% (3×). Incubate the cells in each bath for 10 min.
Transfer the cover slip, cells facing up, to a glass culture dish (see Note 28) filled with 1:1 mix of 100% ethanol and propylene oxide. Infiltrate for 30 min.
Transfer the cover slip, cells facing up, to a glass culture dish (see Note 28) filled with 1:1 mix of embedding resin/propylene oxide. Infiltrate overnight, leaving the culture dish uncovered.
Immediately prior to use, prepare fresh embedding resin and remove air bubbles by placing in a vacuum oven set at 20 °C and with the vacuum turned on. Stop process when resin stops bubbling (about 1 h).
Transfer the cover slip, cells facing up, to an uncovered glass culture dish filled with embedding resin. Infiltrate for 2 h to overnight in a vacuum oven set at 20 °C and with the vacuum turned on.
Cut off the lid of a BEEM capsule, fill the capsule completely with fresh resin, and place the capsule in a BEEM capsule holder.
Cells face the resin-filled BEEM capsule, position the cover slip at a 45° angle over the BEEM capsule, and slowly decrease this angle until the cover slip lays flat on top the resin. Be careful not to create bubbles between the surface of the resin in the BEEM capsule and the cover slip.
Carefully transfer BEEM capsule holder containing the resin-filled BEEM capsule, opening facing up and with cover slip on top, to an oven set up at 60 °C and cure the resin for 48 h (vacuum should be turned off).
Peel off the Thermanox plastic cover slip from the capsule (see Note 29). The cells are now in the resin at the surface of the block that was in contact with the cover slip.
Remove the resin “block” from the BEEM capsule with a BEEM capsule press (see Note 30). Be careful not to damage the block surface holding the cells.
If cross-sectional images of cells are required, use a jeweler saw to saw off a small oblong block (approximately 3 mm wide by 5 mm long) from the end of the block that contains the cells. Mark the sawn surface opposite to the cells with a colored permanent marker. Place this block in a well of a flat plastic embedding mold, with the side with the cells (opposite to the marker mark) (see Note 31) laying flat on the bottom surface of the well and the long axis of the oblong piece parallel to the long axis of the well. Re-embed by filling the well with fresh resin prepared as described in step 4 above and by curing the resin as described in step 8 above.
Mount the block holding the cells in a block trimmer and under a binocular dissecting microscope trim the block edges with a single-edge razor blade to create a four-sided pyramid with walls at a 45° angle and a 0.5–0.75 mm square top surface which contains the cells. Be careful not to trim off the side onto which the cells are embedded.
Mount the trimmed block on the sample holder of the microtone.
Mount the diamond knife on the microtone.
Align the bottom edge of the trapezoidal block so that it parallel to the knife edge.
Fill the diamond knife boat trough with distilled water so it is level with the cutting edge but does not form a meniscus.
Generate ultrathin sections (50–70 nm thickness) (see Note 32). Since the cells are at the surface of the block, it is essential that the very first ultrathin sections generated from the block face are collected and that semi-thin sections (approximately 0.5 μm thickness) are not generated.
Collect ultra-thin sections by holding a copper grid (see Note 14) with a pair of anti-capillary tweezers under the section floating on the water in the knife boat and when the section is positioned over the grid, gently raise the grid to collect the section
Blot the jaws of the tweezers and the bottom of the grid with Whatman #1 filter paper to absorb all of the water and air-dry the grid by placing it, section side up, on the bottom of a clean Petri dish.
Place a 50 μL drop of uranyl acetate staining solution on a piece of Parafilm (see Notes 33 and 34). Gently place the grid section side down, on top of the drop. Stain for 30 min protected from light.
Wash off excess stain by gently placing the grid, section side down, on top of a 50 μL drop of distilled water laid on a piece of Parafilm. Wash for 5 min. Repeat three times.
Place a 50 μL drop of freshly prepared Reynold’s lead citrate staining solution on a piece of Parafilm and arrange a few dry sodium hydroxide pellets around the drop (see Note 35).
Gently place the grid, section side down, on top of the lead citrate drop and cover with a small Petri dish lid or beaker. Stain for 2 min.
Wash off excess stain by gently placing the grid, section side down, on top of a 50 μL drop of distilled water laid on a piece of Parafilm. Wash for 5 min. Repeat three times.
Air-dry the grid by placing it flat, section side up, at the bottom of a clean Petri dish covered with a lid to protect the sections from dust. Dried grids can be stored indefinitely in a grid storage box kept in a dust-free, dry place.
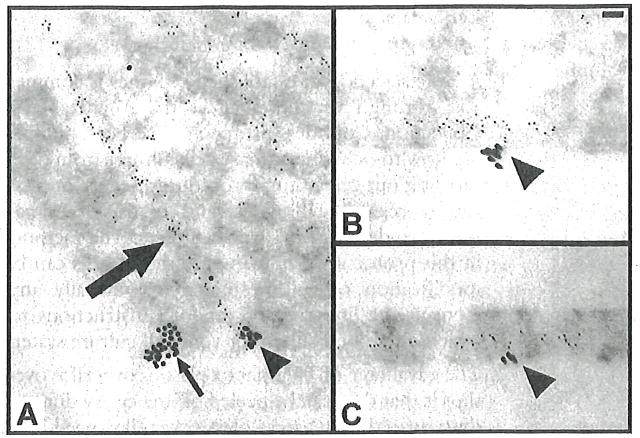
View grids with a TEM. A pre-embedding double-label immunogold electron micrograph of a cultured endothelial cell cross sectioned along the cell-substratum interface is presented in Fig. 1 [9]. To determine whether vimentin intermediate filaments attach to cell-substratum attachment sites containing the matrix receptor β3 integrin, cultured endothelial cells were incubated with a mouse monoclonal anti-vimentin and a rabbit anti-β3 integrin primary antibody mixture and the primary antibodies were visualized with 6 nm gold-conjugated goat anti-mouse IgG and 18 nm gold-conjugated goat anti-rabbit IgG [9]. The double-immunolabeled cells were embedded and sections parallel (A) or perpendicular (B and C) to the substratum were obtained. Uranyl acetate and lead citrate staining was not performed in order to make the spatial relationship of the large and small gold particles more obvious. The 18 nm gold particles labeling β3 integrin can be seen as clusters located at sites where the plasma membrane is in contact with the substratum. The 5 nm gold particles labeling vimentin form linear arrays in the cytoplasm indicative of the filamentous nature of vimentin (Fig. 1a, large arrow). In addition, the close proximity of the 18 nm gold particles with some of the 6 nm particles (Fig. 1a–c, arrowheads) indicates that vimentin filaments are anchored in cell substratum attachment sites containing (β3-integrin. Live-cell imaging experiment later demonstrated that this anchorage regulates the size of β-integrin containing focal contact in response to shear stress [13].
Fig. 1.
Cultured endothelial cells grown on cover slips and fixed in 4% paraformaldehyde in PBS for 5 min followed by extraction in 0.1 % Triton-X-100 for 5 min were double labeled by indirect immunogold using a combination of mouse monoclonal antibodies against vimentin (Sigma-Aldrich) and a rabbit antiserum against (β3 integrin (Chemicon International, Inc., Temecula, CA). Primary antibodies were visualized with 5 nm gold-conjugated goat anti-mouse IgG and 18 nm gold-conjugated goat anti-rabbit IgG secondary antibodies (Jackson ImmunoResearch Labs). Following processing and curing in resin, cells were re-embedded so that cross sections parallel (a) or perpendicular (b and c) could be generated. In a, one group of 18 nm particles (arrowhead) along cell-substratum attached surface of the cell is associated with a, linear array of small gold particles suggestive of a vimentin filament (large arrow) while a second group of 18 nm gold particles is not (small arrow). In b and c, groups of 18 nm particles are associated with linear arrays of 6 nm gold particles along regions of cell-substratum interaction (arrowheads). Note that the sections were not stained with uranyl acetate and Reynold’s lead in order to facilitate the visualization of the 6 nm gold particles. Bar=60 nm (reproduced from [9] with permission of the publisher)
Acknowledgments
Drs. Skalli, Estraño, and Schwartzbach are supported by in-house grants from the University of Memphis. J.C.R. Jones research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Number RO1 AR054184; the content is solely the responsibility of the author and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
A large selection of established cell lines is commercially available through vendors such as the American Type Culture Collection. Countless protocols have been published describing how to establish primary cell cultures from organs. Prior to carrying out cell culture procedures, secure necessary authorizations regarding biosafety and/or use of animal or human tissues to derive primary cell cultures. The methods presented in this protocol for human endothelial cells can be used with modification of culture media for virtually any cell type. Consult the literature or vendor’s instructions to determine which one is best suited for your cell culture system.
The advantage of Thermanox plastic cover slips over glass cover slips is that they can be peeled off the epoxy during the embedding procedure whereas glass cover slips need to be shattered with liquid nitrogen or dissolved with hydrofluoric acid, procedures which may damage the cells embedded in the epoxy. However, if using glass cover slips, ensure that they are not too thick (#1 or #1.5 is fine); otherwise they may not shatter easily with liquid nitrogen during the embedding procedure.
All solutions should be prepared with distilled water and stored at 4 °C unless otherwise stated.
Paraformaldehyde, glutaraldehyde, and osmium tetroxide are toxic and volatile. All work with these chemicals should be performed in a fume hood using gloves and protective clothing. Handling and waste disposal should be done according to the guidelines of the local authorities.
Acetylated BSA or BSA-c (Electron Microscopy Science, Hatfield, PA) at a final concentration of 0.2% can be used in lieu of BSA in the dilution buffer for the primary and secondary antibodies and may be more efficient than BSA at reducing nonspecific immunogold staining.
Preparation of cells for immunolabeling denatures cellular proteins. The antibodies used must thus be able to recognize epitopes on denatured proteins. Only use a primary antibody whose specificity and ability to recognize denatured proteins with high specificity have been verified by Western blotting and fluorescent immunolabeling of cells fixed with 4% paraformaldehyde.
Direct labeling using primary antibodies conjugated with different size gold particles can be used in place of the indirect labeling procedure (i.e., using gold-conjugated secondary antibodies for detection), outlined here. One advantage in doing so is that gold particles attached to the primary antibody are closer to the site of antigen than in the indirect procedure where gold particles are separated from the antigen by both primary and secondary antibody molecules. Nonetheless, our preference is the indirect protocol, since multiple secondary antibodies binding to a single primary antibody result in amplification of the gold label signal output.
This reagent is not needed if performing direct labeling.
The 2 % OsO4 solution can be stored at 4 °C for several months provided that it is kept in the dark. Discard the solution if a black precipitate forms. OsO4 fumes are harmful and all procedures involving OsO4 should be done in a fume hood and OsO4 wastes should be disposed of in closed containers.
To enhance membrane visualization, a 1.5% aqueous solution of potassium ferricyanide (K3Fe(CN)6) can be added to the 1 % OsO4 fixation solution [14]. The mixing of these two solutions should be performed immediately prior to use and the fixation should be undertaken at 4 °C for 4 h in the dark.
UA is toxic and radioactive. Due caution should be taken with its handling. Long-term exposure, skin contact, and inhalation should be avoided.
Embedding medium based on Epon 812 was routinely used until the 1970s. However, in 1978 Epon 812 was discontinued. Nonetheless, a number of companies subsequently developed Epon 812 equivalents which are now used in embedding medium formulations. We indicate one such example.
Unused epoxy resin embedding mix can be stored for up to 6 months at −20 °C but it is preferable to use a freshly prepared mix. Epoxy resin waste should be collected and allowed to polymerize before disposal.
TEM grids are available in different mesh sizes. The smaller the mesh size, the better the sections will be supported by the grids and able to resist tearing when exposed to the electron beam. However, too small a mesh size can result in parts of the section being obscured. With larger mesh sizes, such as the commonly used 200, the sections are more likely to tear under the electron beam. To help preventing this shortcoming, grids coated with a formvar/carbon film can be used. However the formvar/carbon film will sometimes reduce image contrast. Therefore, decision on what mesh size to use and whether or not to use grids coated with a formvar/carbon film is mostly a matter of personal preference.
Carry out the procedures described in this section by using sterile techniques and working in a laminar flow biosafety cabinet.
To determine cell concentration with a hemocytomer, slowly pipet 20 μL of cell suspension under the edge of the cover slip sitting above the hemocytometer gridded area. Then, place the hemocytometer on the stage of a light microscope and count the cells in the central gridded square. The cell concentration (cells/mL) is calculated with the formula (total cells counted × dilution factor × l04). Cell concentrations can also be determined with automated cell counters, but they are more expensive than hemocytometers.
The purpose of this step is to create a “handle” which can be used to carry the cover slip from one solution to another during the immunogold staining. The tweezers need to be sterile for this procedure and this is achieved either by autoclaving or by, dipping the tip of the tweezers in 95 % ethanol and flaming it
The cover slip may float due to air bubbles forming at the bottom of the well when the dish is placed in the incubator. If this happens, remove the dish from the incubator and with sterile tweezers gently push the cover slip down toward the bottom of the dish to dislodge the bubbles.
If the cells are too sparse, it will be difficult to find them when performing TEM observations.
Procedures in this section are carried out at room temperature and do not need to be performed under sterile conditions unless otherwise noted.
Cell fixation that may be optimal for immunofluorescence, such as fixation/extraction in −20 °C acetone or a mix of acetone and methanol, does not preserve high-quality ultrastructure. However, antigenicity may not be preserved in cells fixed with standard TEM fixatives such as 2.5% glutaraldehyde. Thus, compromises must be made between preserving antigen-binding sites while maximizing the preservation of cellular architecture structure. Compromises include fixation in 4% paraformaldehyde or 4% paraformaldehyde plus 0.1–0.2% glutaraldehyde. Immunofluorescence can be used to test whether or not the aldehyde fixation solution preserves the antigenicity of the protein being labeled.
Alternatively, the specimen can be incubated in 0.5 mg/mL sodium borohydride (NaBH4) in PBS for 20 min to reduce the free aldehyde groups that can nonspecifically bind colloidal gold.
Permeabilization is not needed for extracellular epitopes. Increasing permeabilization time and/or detergent concentration will result in degradation of cellular membrane and membranous organelle ultrastructure. Some protocols employ 0.1 % NP40 in PBS in lieu of Triton-X-100.
A 24-well plate is used at this stage to reduce cost by minimizing the amount of antibody used.
It is useful to start with the dilution of primary antibody that gives good consistent staining in sections prepared for immunofluorescence microscopy. To reduce nonspecific binding of the secondary antibody to the specimen, serum of the animal species in which the secondary antibody is made should be added to the primary antibody diluent at a dilution of 1:20–1:100. To determine the extent of nonspecific immunogold labeling, experiments should be performed by omitting the primary antibody. Additional controls may also involve using a primary antibody known not to recognize proteins of the species from which the specimen was taken.
This step is not needed if performing direct labeling (see Note 7).
PBS and TBS are optimal for antibody binding, but may form precipitates with uranyl acetate, osmium, and/or lead citrate and thus must be replaced by EM wash buffer at this stage. EM wash buffer is not used to dilute antibodies in previous steps because it is not optimal for antibody binding to antigen. An alternative to EM wash buffer is 0.1 M phosphate buffer, pH 7.2.
Propylene oxide will dissolve most plastic dishes. The plastic of Thermanox cover slips, however, is not dissolved by propylene oxide.
If the cells were grown on glass cover slips, the glass cover slips can be removed (shattered) from the BEEM capsule by immersion in liquid nitrogen or dissolved with hydrofluoric acid.
Alternatively, slice the sides of the BEEM capsule with a razor blade to loosen the block but this carries a. higher risk to damage the block.
The fixed cells may also be visible as a black or brown line using a binocular dissecting microscope. This line can also be used for orientation.
50–70 nm Ultra-thin sections will appear to have a silvery shine when floating on the surface of the water and viewed with the binocular mounted on the ultramicrotome.
Sections can be dried and viewed with the transmission electron microscope prior to staining with heavy metal salts (uranyl acetate and lead citrate) to determine whether the immunolabeling was efficient. Gold particles, especially small ones (5 nm), may be more readily visualized in the absence of heavy metal staining and in that case heavy metal staining may be omitted. However, biological structures may be difficult to identify in the absence of heavy metal staining.
Uranyl acetate staining enhances the electron contrast of biological structures by interacting with lipids and proteins. However, needle-like crystal precipitates may be present in uranyl acetate solutions that have not been cleared by filtration or centrifugation or may form in the presence of residual PBS on the section.
Lead citrate staining enhances the electron contrast of biological structures by interacting with proteins and glycogens. Lead citrate, however, forms precipitates of lead carbonate in the presence of CO2. The sodium hydroxide pellets adsorb the CO2 preventing precipitate formation.
References
- 1.Faulk WP, Taylor GM. An immuno-colloid method for the electron microscope. Immunochemistry. 1971;8:1081–1083. doi: 10.1016/0019-2791(71)90496-4. [DOI] [PubMed] [Google Scholar]
- 2.Frens G. Controlled nucleation for the regulation of the particle size in monodisperse gold suspensions. Nature. 1973;241:20–23. [Google Scholar]
- 3.Psarra AM, Solakidi S, Sekeris CE. The mitochondrion as a primary site of action of steroid and thyroid hormones: presence and action of steroid and thyroid hormone receptors in mitochondria of animal cells. Mol Cell Endocrinol. 2006;246:21–33. doi: 10.1016/j.mce.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 4.Frotscher M, Studer D, Graber W, Chai X, Nestel S, Zhao S. Fine structure of synapses on dendritic spines. Front Neuroanat. 2014;8:94. doi: 10.3389/fnana.2014.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bendayan M, Shore GC. Immunocytochemical localization of mitochondrial proteins in the rat hepatocyte. J Histochem Cytochem. 1982;30:139–147. doi: 10.1177/30.2.7061817. [DOI] [PubMed] [Google Scholar]
- 6.Skalli O, Jones JC, Gagescu R, Goldman RD. IFAP 300 is common to desmosomes and hemidesmosomes and is a possible linker of intermediate filaments to these junctions. J Cell Biol. 1994;125:159–170. doi: 10.1083/jcb.125.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones L, Rogers G, Rufaut N, Sinclair R. Location of keratin-associated proteins in developing fiber cuticle cells using immunoelectron microscopy. Int J Trichology. 2010;2:89–95. doi: 10.4103/0974-7753.77512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klatte DH, Kurpakus MA, Grelling KA, Jones JCR. Immunochemical characterization of hemidesmosomal components and their expression in cultured epithelial cells. J Cell Biol. 1989;104:3377–3390. doi: 10.1083/jcb.109.6.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gonzales M, Weksler B, Hopkinson SB, Tsuruta D, Goldman RD, Yoon KJ, Flitney FW, Jones JCR. Structure and function of a vimentin-associated matrix adhesion in endothelial cells. Mol Biol Cell. 2001;12:85–100. doi: 10.1091/mbc.12.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibson KH, Vorkel D, Meissner J, Verbavatz JM. Fluorescing the electron: strategies in correlative experimental design. Methods Cell Biol. 2014;124:23–54. doi: 10.1016/B978-0-12-801075-4.00002-1. [DOI] [PubMed] [Google Scholar]
- 11.Redemann S, Müller-Reichert T. Correlative light and electron microscopy for the analysis of cell division. J Microsc. 2013;251:109–112. doi: 10.1111/jmi.12056. [DOI] [PubMed] [Google Scholar]
- 12.Melo RC, Morgan E, Monahan-Earley R, Dvorak AM, Weller PF. Pre-embedding immunogold labeling to optimize protein localization at subcellular compartments and membrane microdomains of leukocytes. Nat Protoc. 2014;9:2382–2394. doi: 10.1038/nprot.2014.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsuruta D, Jones JC. The vimentin cytoskeleton regulates focal contact size and adhesion of endothelial cells subjected to shear stress. J Cell Sci. 2003;116:4977–4984. doi: 10.1242/jcs.00823. [DOI] [PubMed] [Google Scholar]
- 14.Rivlin PK, Raymond PA. Use of osmium tetroxide-potassium ferricyanide in reconstructing cells from serial ultrathin sections. J Neurosci Methods. 1986;20:23–33. doi: 10.1016/0165-0270(87)90036-7. [DOI] [PubMed] [Google Scholar]