Figure 5.
Differential Regulation of Transcription by SNHht and Pam3
(A–F) Microarray analysis performed on M-CSF BMDMs differentiated from 5 WT mice and stimulated for 3 hr.
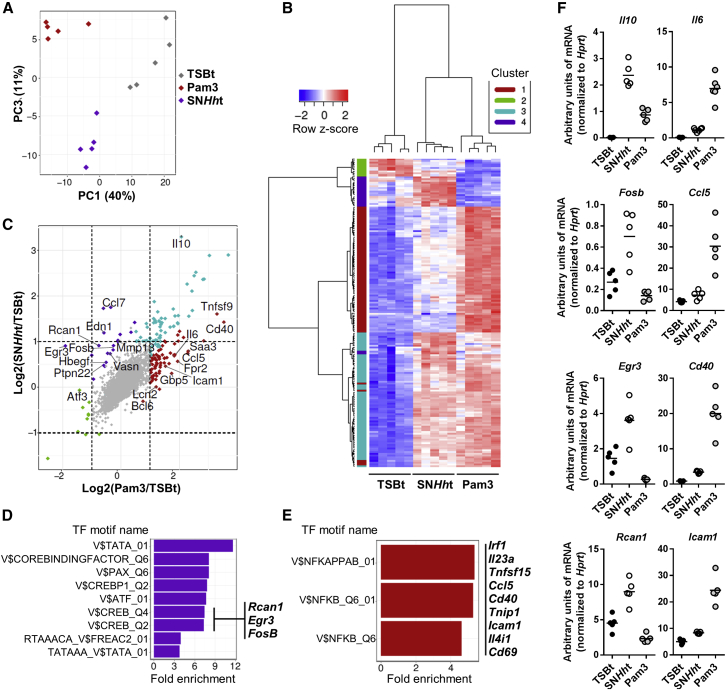
(A) Principle components analysis (PCA) of transcriptional profiles (empirical Bayes normalized expression values from LIMMA) across three conditions (TSBt, Pam3, and SNHht, n = 5/condition, 16,692 probes).
(B) Hierarchical clustering heatmap (Manhattan distance with Ward clustering of empirical Bayes normalized expression values) of the union of differentially expressed probes between any condition-condition contrast (i.e., TSBt versus Pam3, TSBt versus SNHht, or Pam3 versus SNHht) (n = 172 probes, n = 146 genes). Probes were assigned to clusters using k-means clustering (k = 4) and cluster assignments are annotated as colors to the left of the heatmap. The number of probes assigned to each cluster were: cluster 1 = 75, cluster 2 = 10, cluster 3 = 68, and cluster 4 = 19.
(C) Scatterplot displaying the relationship between log2 (fold changes) for each gene shown in (B) obtained when comparing either Pam3 (x axis) or SNHht (y axis) with the TSBt control condition. Colors represent the cluster assignments.
(D and E) Transcription factor (TF) motif enrichment analysis among genes assigned to (D) cluster 4 (i.e., specifically induced by SNHht) and (D) cluster 1 (i.e., specifically induced by Pam3). Motif names correspond to Transfac motif identifiers derived from the Molecular Signatures Database (MSigDB). Motifs significantly overrepresented at an adjusted p < 0.05 (permutation test) are shown.
(F) mRNA levels of CREB (Il10, Fosb, Egr3, Rcan1) and NF-κB (Il6, Ccl5, Cd40, Icam1) target genes were validated by qRT-PCR.