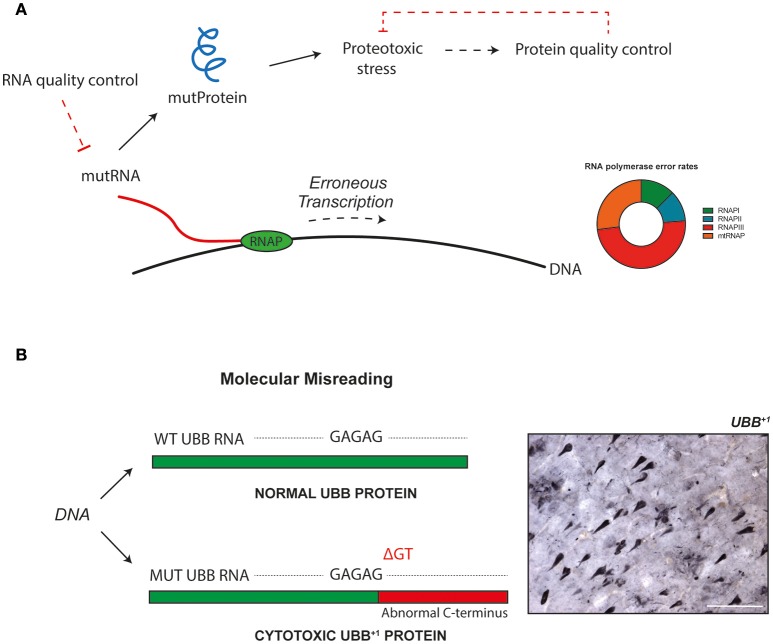
Figure 1.
Erroneous gene transcription can bring about abnormal proteins with cytotoxic properties. These molecules impact cellular function, and may be implicated in aging and disease. Dirty transcripts yield toxic proteins. (A) Transcriptional misreading, i.e., the inaccurate conversion of DNA to RNA by RNA polymerase (RNAP), results in mutant transcripts that can be translated into abnormal proteins. These mutant proteins induce proteotoxic stress in cells and impact cellular physiology. Cells use several mechanisms for quality control of gene expression, to prevent abnormal molecules from being generated and to accumulate. In addition to the proofreading abilities of polymerase enzymes, specific mechanisms exist to control fidelity beyond synthesis. For example, mRNA surveillance pathways ensure mRNA quality (Doma and Parker, 2007), and abnormal proteins can be degraded by an assembly of chaperons, the ubiquitin-proteasome system and autophagy (Goldberg, 2003). It has been suggested that mutations in synthesis machinery could result in a positive feedback loop, corresponding to an error catastrophe (Martin and Bressler, 2000). Impaired quality control mechanisms, perhaps the result of errors during synthesis of their constituents, could also contribute to such a phenomenon. (B) Molecular misreading has been found to introduce sequence differences into nascent transcripts and generates frame-shifted proteins with cytotoxic properties. Ubiquitin-B+1 (UBB+1) is an example of such a protein and accumulates in the neuropathological hallmarks of Alzheimer's disease. It has been shown that when the levels of proteotoxic stress induced by UBB+1 surpass cellular redundancy, this will lead to pathogenicity. The insert shows a section from an Alzheimer's disease patient brain, containing several UBB+1-positive structures. Bar = 100 μm.