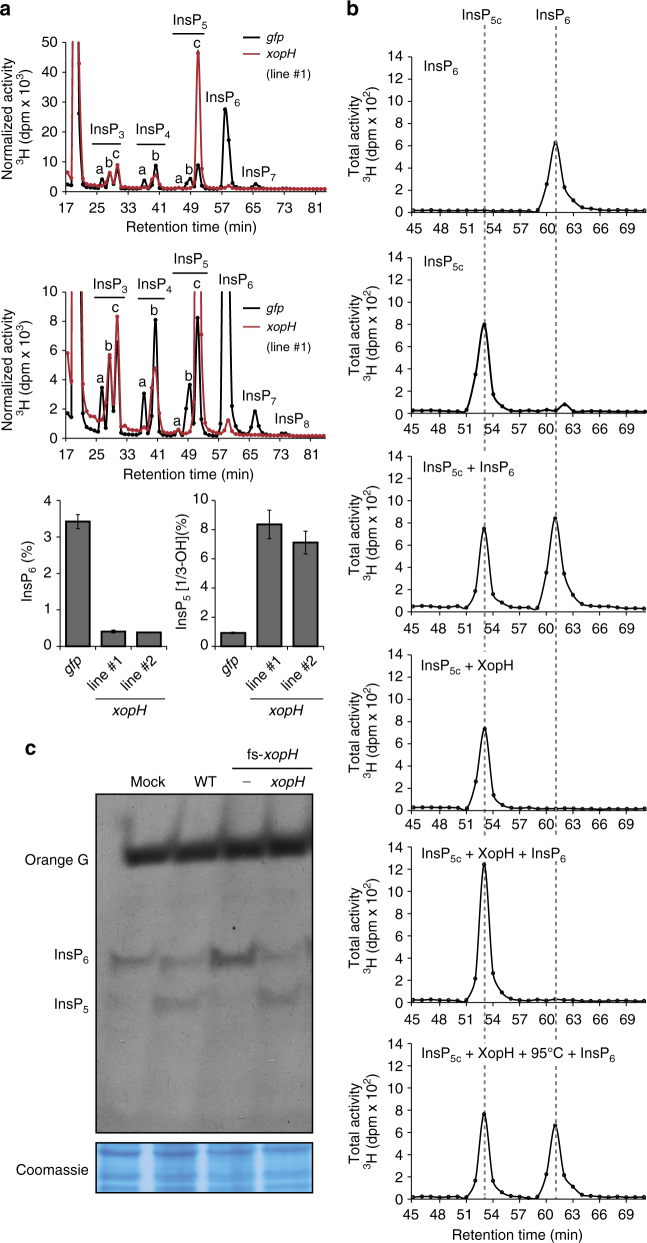
Fig. 7.
XopH causes InsP5 accumulation in planta. a Upper panel: HPLC profiles of extracts from transgenic N. benthamiana leaves, labeled with [3H]-myo-inositol. Based on published chromatographic mobilities69, InsP5a represents InsP5 [2-OH], InsP5b represents InsP5 [4/6-OH], and InsP5c represents InsP5 [1/3-OH]. The isomeric natures of InsP3a-c, InsP4a-b, InsP7, and InsP8 are unknown. Middle panel: Zoom-in on the HPLC profiles. Lower panels: Relative amounts of InsP6 and InsP5c in N. benthamiana expressing gfp and two independent lines expressing xopH. Error bars indicate s.e.m. b Digestion and HPLC analyses of plant-purified InsPx species. InsP5c and InsP6 were purified from [3H]-myo-inositol-labeled xopH- and gfp-expressing N. benthamiana seedlings, respectively (see “Methods” section). XopH-treated or non-treated InsPx species (as designated) were then separated by SAX-HPLC. XopH was inactivated by incubating the reaction mixture at 95 °C for 15 min. c XopH hydrolyzes InsP6 to InsP5 during Xcv infection. TiO2 bead enrichment of inositol polyphosphates from N. benthamiana leaf extracts infected with Xcv 85-10ΔxopQ (WT), 85-10ΔxopQ_fs-xopH (fs-xopH), and the complemented mutant, where xopH was re-integrated into the genome (see “Methods” section). Inositol polyphosphates were eluted from TiO2 beads, resolved by PAGE and visualized by toluidine blue staining. Protein extracts were visualized by Coomassie blue as a loading control. The experiments were repeated twice (a, c) or once (b) with similar results