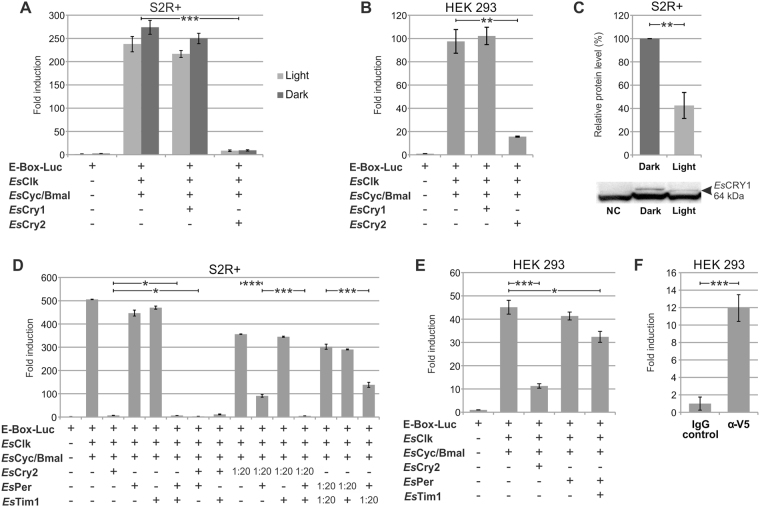
Figure 5.
Functional characterization of the putative EsCLK:EsCYC/BMAL’s inhibitors. (A,B) EsCRY1 and EsCRY2 functional validation by luciferase assay in S2R + and HEK293 cells, respectively. Cells were transfected with indicated constructs. Negative control set as 1. Data are represented as mean ± SEM (n = 3 independent transfections). (C) Western blot and relative quantification of EsCRY1 protein in the dark and after a 8 hours light pulse in Drosophila cells. Data are represented as mean ± SD (n = 3 independent transfections). NC: negative control. (D,E) Comparison of the effectiveness of EsPER, EsTIM1, and EsCRY2 for inhibiting the transcription of the E-box/luciferase reporter mediated by the EsCLK:EsCYC/BMAL dimer in S2R + and HEK293 cells respectively. S2R + and HEK293 cells were transfected with the indicated constructs. Negative control set as 1. Data are represented as mean ± SD (n = 3 independent transfections). (F) Co-immonoprecipitation of EsCRY2 and EsCYC/BMAL quantified by luciferase assay. EsCyc/Bmal C-terminally fused to luciferase (EsCyc/Bmal-LUC) was co-immunoprecipitated with EsCry2-V5 and anti-V5 antidody in HEK293 cells. Data are presented as mean ± SD (n = 3 independent transfections). Student’s t-test Bonferroni-corrected p-values for all the experimental comparisons discussed were presented in Supplementary Table 3. Statistical significance of the most relevant comparisons were shown as *p < 0.05, **p < 0.01, and ***p < 0.005.