Fig. 1.
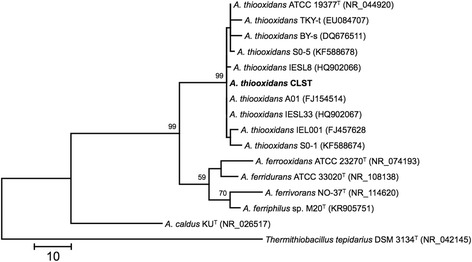
Phylogenetic tree based on the 16S rRNA gene sequences highlighting the position of 10.1601/nm.2199 strain CLST relative to other type and non-type strains of the genus 10.1601/nm.2198. The GenBank database accession codes are indicated between brackets. The evolutionary history was inferred by using the Maximum Parsimony and the Subtree-Pruning-Regrafting (SPR) algorithm with search level 1 [52]. The initial trees were obtained by the random addition of sequences. The analysis involved 16 nucleotide sequences and a total of 1307 non-ambiguous positions in the final dataset. Evolutionary analyses were conducted in MEGA version 6.22 [53]. Tree construction used a bootstrapping process repeated 1000 times to generate a majority consensus tree. A sequence from 10.1601/nm.2205 was used as outgroup. The tree is drawn to scale, with branch lengths calculated using the average pathway method [53]; the scale bar corresponds to the number of changes over the whole sequence
